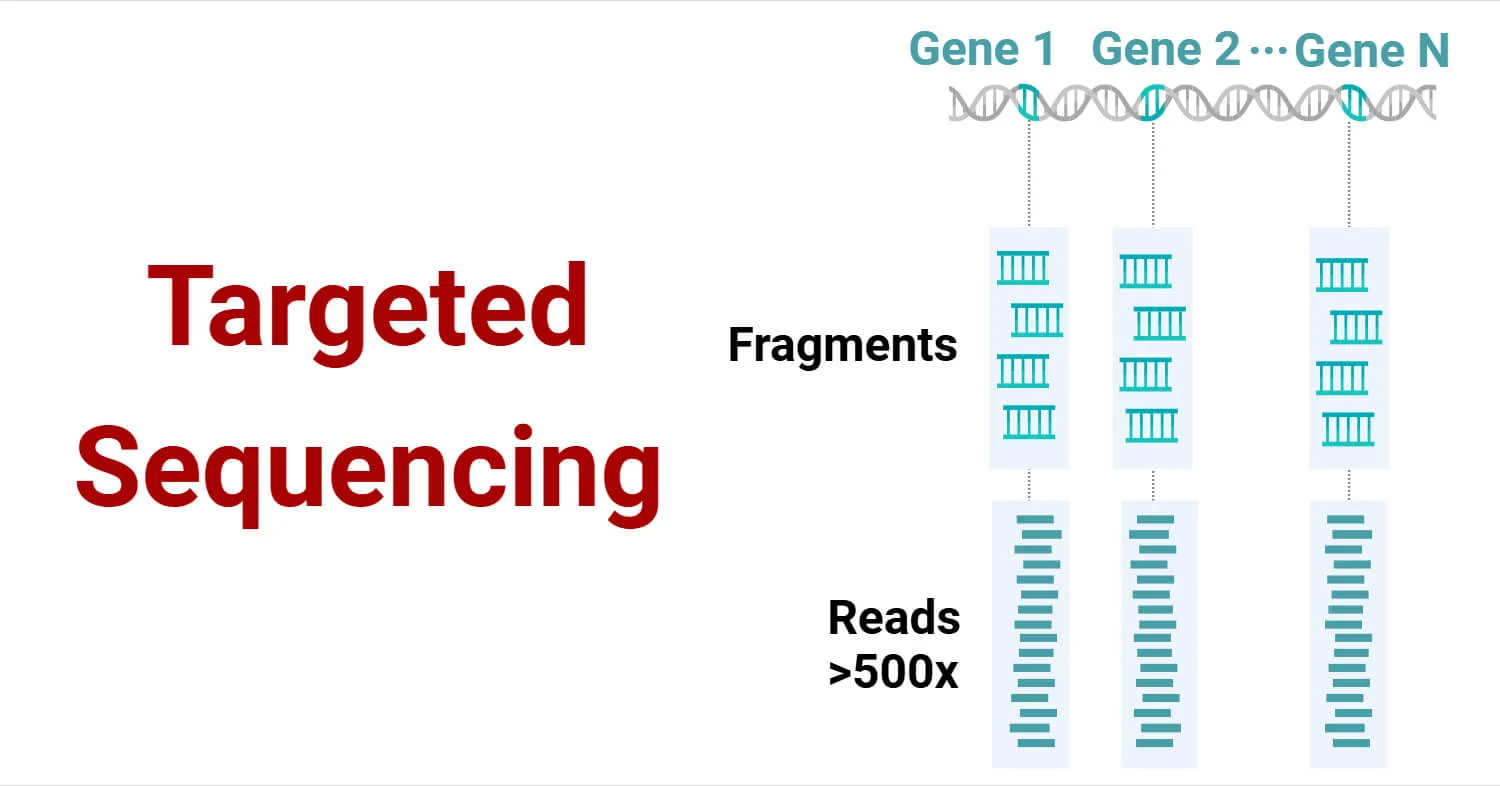
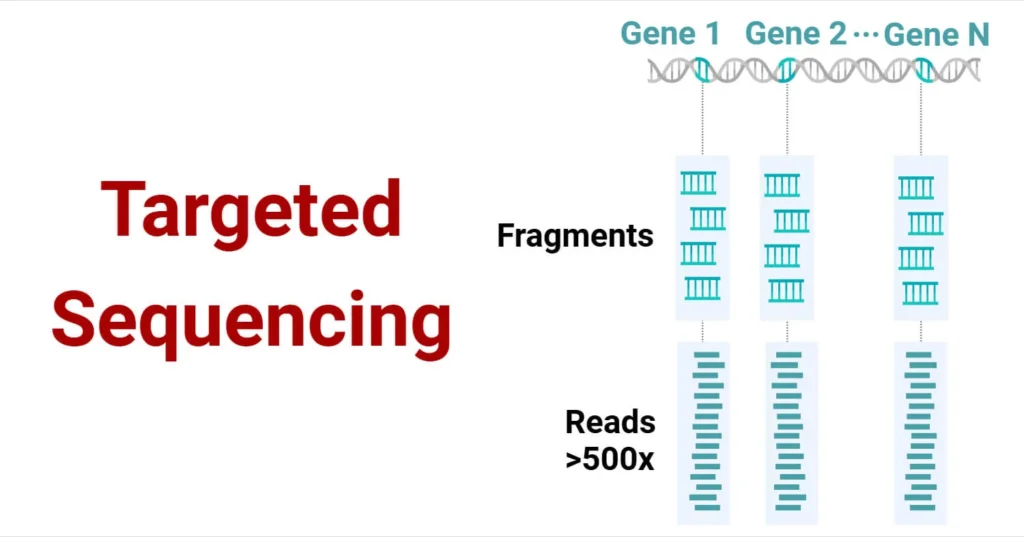
Targeted sequencing is a method of sequencing specific genomic regions of interest rather than sequencing the entire genome. This approach is highly effective for analyzing particular genes or regions, offering a more efficient and cost-effective alternative to whole genome sequencing (WGS). By focusing on specific genomic areas, targeted sequencing provides higher depth coverage, which improves the detection of genetic variants, including rare ones.
Principle of Targeted Sequencing
The principle behind targeted sequencing revolves around focusing on specific genomic regions for sequencing. Rather than sequencing the entire genome, targeted sequencing uses specialized techniques to enrich and capture the regions of interest. This enriched DNA is then sequenced for a more detailed and efficient genetic analysis. The method significantly enhances sequencing depth, increasing the likelihood of detecting genetic variants, especially in samples with degraded DNA.
Target enrichment can be accomplished in two primary ways: using probes that specifically capture DNA sequences of interest or primers that amplify the regions of focus. This focused approach optimizes sequencing resources and provides more accurate data from the selected regions.
Process/Steps of Targeted Sequencing
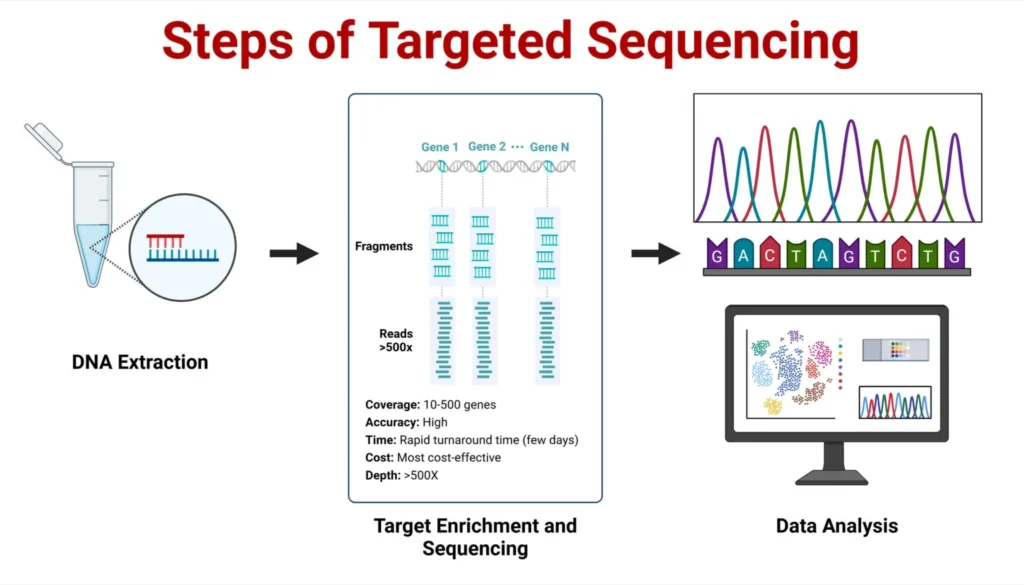
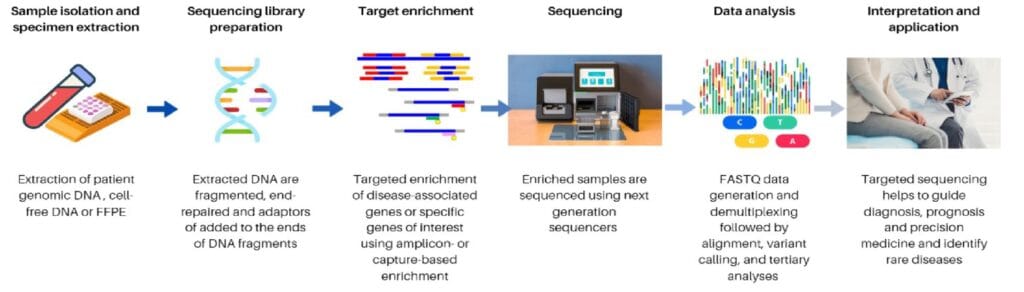
- Sample Preparation
High-quality DNA or RNA is extracted from biological samples. The quality of the nucleic acids is crucial as it impacts all subsequent steps. Contamination or degradation can affect the sequencing results. - Library Preparation
The isolated DNA is fragmented into smaller pieces. This is followed by end repair and the addition of sequencing adapters. This ensures the DNA fragments are ready for target enrichment and sequencing. - Target Enrichment
This step is where targeted sequencing distinguishes itself from whole genome sequencing. The DNA regions of interest are selectively enriched using either hybridization-based capture or amplicon-based enrichment.- Hybridization-Based Capture: Probes are used to bind to the DNA regions of interest, and streptavidin-coated magnetic beads separate the captured DNA.
- Amplicon-Based Enrichment: Primers amplify the target regions using PCR, increasing the concentration of DNA in the regions of interest.
- Sequencing
After enrichment, the DNA library is sequenced using next-generation sequencing (NGS) platforms such as Illumina. The sequencing process generates millions of short reads that cover the enriched regions of the genome. - Data Analysis
The sequencing data is aligned to a reference genome, and various bioinformatics tools are employed to analyze the genetic variants. This process involves detecting structural variants, annotating genetic variations, and ensuring high-quality sequencing reads.
Methods of Target Enrichment
- Hybridization-Based Capture:
Biotinylated oligonucleotide probes bind to the target DNA regions, and the captured DNA is separated using magnetic beads. Unique Molecular Identifiers (UMIs) may also be included to reduce errors and biases in PCR. - Amplicon-Based Enrichment:
This method uses primers specific to the target regions to amplify the genomic DNA via PCR. It is faster and more cost-effective than hybridization-based capture but has limitations in achieving uniform coverage, especially in complex or poor-quality DNA samples.
| Characteristic | Hybridization Capture | Amplicon Sequencing |
|---|---|---|
| Method | Uses probes to capture DNA regions of interest | Amplifies regions using specific primers |
| Complexity and Speed | More complex, slower | Faster and simpler |
| Sample Input | Requires more DNA input | Requires less DNA input |
| Cost | More expensive | More cost-effective |
| Coverage Uniformity | More uniform coverage | Less uniform coverage |
| Mismatch Tolerance | Higher tolerance for mismatches | Lower tolerance for mismatches |
| Amplification Errors | Fewer PCR duplicates, accurate alignment | Risk of PCR duplicates, bias in data |
| Sensitivity | Better for low-quality DNA | Prone to off-target amplification |
Advantages of Targeted Sequencing
- Cost-Effective: It is a more affordable option than whole genome sequencing since it sequences only the regions of interest, reducing the overall cost.
- Increased Efficiency: By focusing on specific regions, targeted sequencing avoids wasting resources on irrelevant genomic data, leading to faster and more focused analyses.
- High Coverage: The targeted approach ensures that the regions of interest are sequenced more deeply, which improves the accuracy of variant calling.
- Reduced Data Complexity: With fewer regions sequenced, the resulting data is smaller and more manageable, simplifying data analysis.
Limitations of Targeted Sequencing
- Limited Scope: Since it focuses only on preselected regions, targeted sequencing may miss important variants outside of these regions.
- Incomplete Genome Picture: It doesn’t provide a complete representation of the entire genome, unlike whole genome sequencing.
- Complex Panel Design: Designing sequencing panels to target specific regions can be challenging and time-consuming.
- Coverage Uniformity: Achieving uniform coverage across all targeted regions can be difficult, especially with low-quality or complex samples.
- PCR Bias: Amplicon-based sequencing can introduce PCR amplification errors, leading to biases in data analysis.
Applications of Targeted Sequencing
- Clinical Diagnostics: Targeted sequencing is widely used in diagnosing inherited genetic disorders and identifying disease-causing mutations.
- Cancer Research: By focusing on cancer-related genes, targeted sequencing helps identify mutations and study tumor biology to develop personalized treatments.
- Infectious Diseases: Targeted sequencing is used to study viral genomes and track genetic variations in infectious agents during outbreaks.
- Biomarker Discovery: It is used to identify biomarkers associated with diseases, improving diagnostic and therapeutic strategies.
- Microbial Profiling: Targeted sequencing is useful in metagenomics to profile microbial communities and study the microbiome.
Diagram of Targeted Sequencing Workflow
Targeted sequencing is a powerful tool that provides focused insights into specific genomic regions. It is widely used in clinical diagnostics, cancer research, and microbial profiling due to its high efficiency, cost-effectiveness, and deep coverage of selected regions. Despite its limitations, such as its inability to capture the entire genome and the potential for PCR bias, targeted sequencing remains a preferred method for studies that require in-depth analysis of specific genes or regions.