RNA sequencing (RNA-Seq) is a revolutionary molecular biology technique that allows for the comprehensive analysis of the transcriptome, providing insights into the gene expression levels of a cell. This technology has transformed transcriptomics, enabling scientists to explore the intricacies of cellular processes, study diseases, and accelerate pharmaceutical development. This article delves into the definition, principles, steps, types, applications, advantages, and limitations of RNA sequencing.
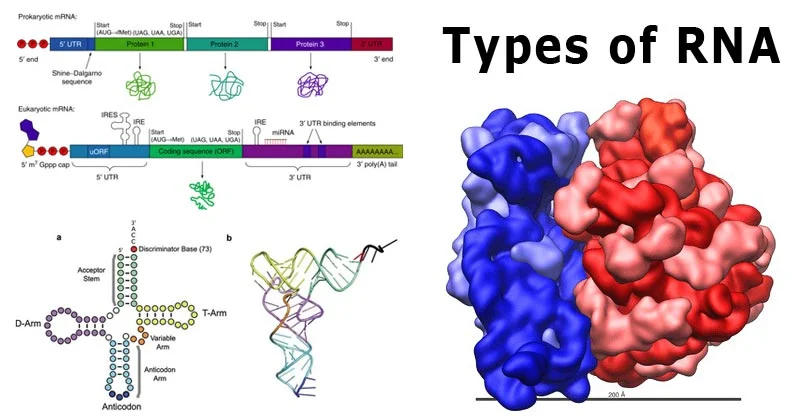
Definition of RNA Sequencing
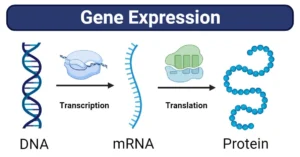
RNA sequencing (RNA-Seq) is a high-throughput technique used to sequence the RNA present in a sample. It provides insights into the entire set of RNA molecules, including both coding and non-coding transcripts. Unlike traditional approaches that focus solely on specific RNA types, RNA-Seq allows researchers to study all RNA species simultaneously, facilitating a deeper understanding of gene expression, alternative splicing, and post-transcriptional modifications.
Principle of RNA Sequencing
RNA sequencing operates on a principle similar to that of DNA sequencing, but the target in RNA-Seq is the RNA molecule. The basic process involves isolating RNA from a sample, converting it into complementary DNA (cDNA), and sequencing the cDNA. Unlike DNA sequencing, RNA-Seq provides a snapshot of gene activity, offering a more dynamic view of cellular processes by measuring the levels of different RNA molecules.
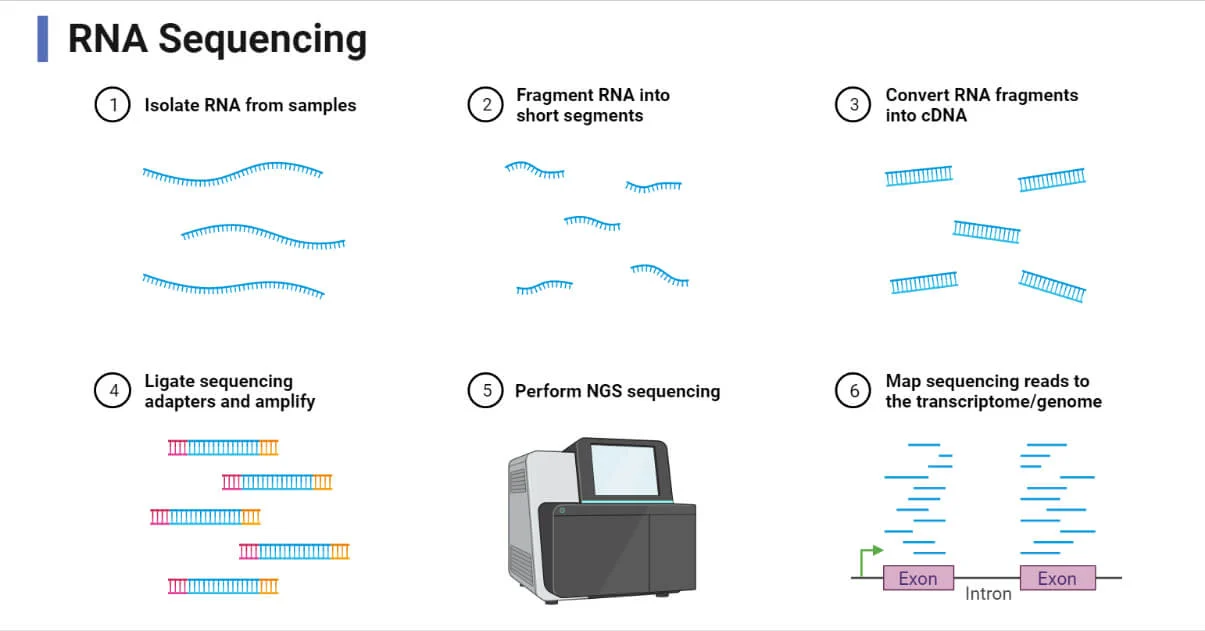
The RNA-Seq process involves several stages:
- RNA Extraction: RNA is extracted from the sample.
- cDNA Synthesis: RNA is reverse-transcribed into cDNA, which is more stable and easier to handle.
- Library Preparation: cDNA is fragmented, adapters are ligated, and the resulting cDNA fragments are amplified.
- Sequencing: The cDNA is sequenced using high-throughput sequencing technologies.
- Data Analysis: The sequence data is analyzed to quantify gene expression, identify mutations, and detect alternative splicing.
Types of RNA Sequencing
RNA sequencing can be categorized based on the method of cDNA formation or the type of RNA being sequenced.
1. Based on the Formation of cDNA
- Direct RNA Sequencing: This method sequences RNA molecules directly without converting them into cDNA. While this avoids biases introduced during cDNA synthesis, RNA molecules are unstable, making this method more challenging to work with.
- Indirect RNA Sequencing (cDNA Sequencing): This is the most common method, where RNA is first converted into cDNA and then sequenced. This method is more stable and easier to work with, especially for complex samples.
2. Based on the Type of RNA Sequenced
- Whole Transcriptome RNA Sequencing (WTS): This approach sequences all RNA types present in a sample, providing a comprehensive view of gene expression, including coding and non-coding RNAs.
- mRNA Sequencing (mRNA-Seq): This method focuses specifically on messenger RNA (mRNA), the RNA molecules that encode proteins. mRNAs are isolated, typically using poly-A tail selection, and sequenced.
- tRNA and rRNA Sequencing: These methods sequence transfer RNA (tRNA) and ribosomal RNA (rRNA), although they are less commonly used due to the abundance of these molecules in cells.
- Targeted RNA Sequencing: This approach targets and sequences specific RNA molecules of interest, often used for studying particular genes or pathways.
- Small RNA Sequencing: Small non-coding RNAs like microRNAs (miRNAs), small interfering RNAs (siRNAs), and piRNAs are sequenced in this method, which helps in studying gene regulation.
- Single-Cell RNA Sequencing: This advanced technique involves sequencing RNA from individual cells, providing insights into cellular heterogeneity and gene expression dynamics at the single-cell level.
Procedure/Steps of RNA Sequencing
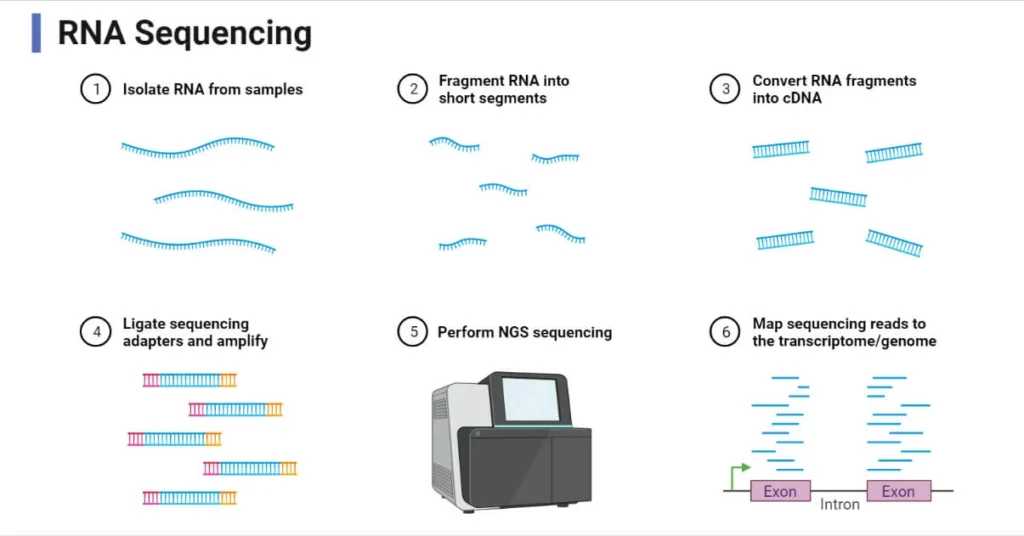
The most widely used RNA-Seq method involves converting RNA into cDNA before sequencing. The general workflow for this method includes the following steps:
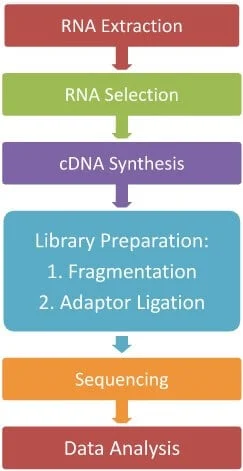
1. RNA Extraction
The first step in RNA-Seq is the extraction of RNA from the sample. This involves lysing the cells to release their RNA and then purifying the RNA using techniques like organic solvent extraction. The RNA is then treated to remove any contaminating DNA.
2. RNA Selection
In RNA-Seq, RNA molecules need to be selected for sequencing. Typically, mRNA is the primary target because it contains the coding information for proteins. mRNA is isolated by selecting for the poly-A tail using techniques like poly-A chromatography or magnetic bead selection. In whole transcriptome sequencing, all RNA types are retained.
3. cDNA Synthesis
RNA molecules are reverse-transcribed into complementary DNA (cDNA) using reverse transcriptase enzymes. This step is crucial because cDNA is more stable and easier to handle than RNA. After synthesizing the first strand of cDNA, the second strand is synthesized using DNA polymerase.
4. Library Preparation
The cDNA is fragmented into smaller pieces and adapter sequences are ligated to both ends of the fragments. These adapters serve as binding sites for the sequencing platform. The cDNA fragments are then amplified using polymerase chain reaction (PCR).
5. Sequencing
The prepared cDNA library is sequenced using next-generation sequencing (NGS) technologies, such as Illumina or PacBio. These platforms generate millions of short sequence reads from the cDNA fragments.
6. Data Analysis
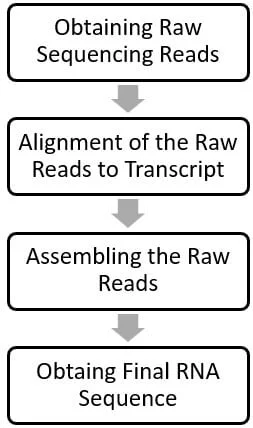
Once sequencing is complete, the resulting raw data is analyzed using bioinformatics tools. This involves aligning the sequence reads to a reference genome or transcriptome, assembling them into full-length transcripts, and quantifying gene expression.
Applications of RNA Sequencing
RNA sequencing has a wide range of applications in various fields, particularly in genomics, medicine, and drug development.
1. Gene Expression Profiling
RNA-Seq provides a detailed and accurate measure of gene expression levels in different samples. It helps to identify which genes are active, and to quantify the amount of RNA produced from each gene.
2. Mutation Detection
RNA-Seq can identify mutations, including single nucleotide polymorphisms (SNPs), insertions, deletions, and gene fusions. This is particularly useful in studying cancer and genetic diseases, where mutations in RNA can result in altered protein production.
3. Alternative Splicing Analysis
RNA-Seq can detect alternative splicing events, where a single gene can produce multiple RNA isoforms, contributing to the complexity of the transcriptome. This is crucial for understanding gene regulation and the functional diversity of proteins.
4. Disease Diagnosis
RNA-Seq can be used for diagnosing genetic diseases, cancer, and other disorders by identifying specific RNA markers associated with disease states. For instance, the presence of mutated mRNAs can be used to diagnose cancers.
5. Drug Development and Pharmacogenomics
RNA sequencing is used to identify potential drug targets, monitor drug effects, and personalize treatments. In pharmacogenomics, RNA-Seq can help understand how different individuals respond to drugs based on their gene expression profiles.
6. Ribosomal Profiling
Ribosomal profiling is a technique that identifies which mRNAs are being actively translated into proteins. This information is valuable for understanding protein synthesis and cellular function.
7. Studying Small RNAs
Small RNA sequencing allows the study of non-coding RNAs like microRNAs (miRNAs) and small interfering RNAs (siRNAs), which play a significant role in gene regulation.
Advantages of RNA Sequencing
RNA sequencing offers several advantages over traditional gene expression analysis methods:
- High Sensitivity: RNA-Seq can detect low-abundance transcripts, providing a more accurate measure of gene expression.
- Quantitative Analysis: RNA-Seq provides both qualitative and quantitative data, allowing for the precise measurement of gene expression levels.
- No Need for Predefined Probes: Unlike microarrays, RNA-Seq does not require predefined probes, making it suitable for studying both known and unknown RNA species.
- Comprehensive: RNA-Seq can simultaneously analyze all RNA types in a sample, providing a comprehensive view of the transcriptome.
Limitations of RNA Sequencing
Despite its advantages, RNA sequencing has some limitations:
- High Cost: RNA-Seq can be expensive due to the need for specialized equipment and reagents.
- Time-Consuming: The process of library preparation, sequencing, and data analysis can be time-intensive.
- Data Complexity: Analyzing RNA-Seq data requires significant computational resources and expertise, as it involves handling large volumes of data.
- RNA Degradation: RNA is less stable than DNA, making it challenging to work with and increasing the chances of degradation during the extraction and sequencing process.
Common RNA Sequencing Platforms
Several platforms are available for RNA-Seq, each with its own advantages and applications:
- Illumina HiSeq Platform: One of the most widely used platforms for RNA-Seq, offering high throughput and accuracy.
- PacBio SMRT Sequencing: Known for long-read sequencing, which can help with more accurate assembly of complex transcriptomes.
- Oxford Nanopore: A portable platform that allows for real-time sequencing, offering long reads for more comprehensive transcript analysis.
RNA sequencing has become an indispensable tool for genomics and molecular biology research. Its ability to profile the entire transcriptome has provided valuable insights into gene expression, disease mechanisms, and drug development. While it offers numerous advantages, including high sensitivity and comprehensive data, the challenges of cost, complexity, and data analysis need to be addressed for its broader adoption. As sequencing technologies continue to evolve, RNA-Seq will play a critical role in advancing our understanding of biology and medicine.