Biomedical Importance
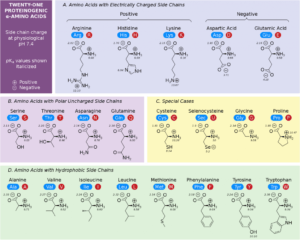
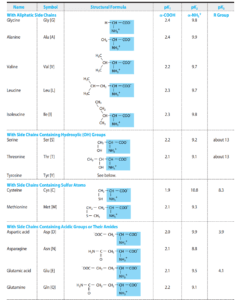
Proteins are the central working molecules of life, performing enzymatic, structural, regulatory, and signaling roles. Knowing the primary structure—the linear sequence of amino acids—is crucial because it dictates how a protein folds and functions. Determining primary structure is fundamental in understanding disease mechanisms, designing drugs, producing recombinant proteins, and characterizing post-translational modifications.
Proteins & Peptides Must Be Purified Prior to Analysis
Accurate sequence determination requires highly purified proteins or peptides. Impurities can obscure analytical results, produce misleading peaks in chromatograms, or interfere with enzymatic or chemical sequencing reactions. Therefore, purification is the first and indispensable step.
Chromatographic Methods for Purification
- Column Chromatography:
The backbone of protein purification. A sample is applied to a column packed with a stationary phase; proteins separate based on their interactions with this phase as buffer flows through.

- Partition Chromatography:
Separates proteins based on differences in solubility between two immiscible phases (liquid–liquid or liquid–solid). - Size-Exclusion Chromatography (Gel Filtration):
Separates proteins by size; larger molecules elute first because they are excluded from the pores of the stationary phase. - Adsorption Chromatography:
Exploits selective adsorption of proteins to solid surfaces. Binding and elution are controlled by changing pH, ionic strength, or solvents. - Ion-Exchange Chromatography:
Uses charged resins to separate proteins according to their net charge at a given pH. - Hydrophobic Interaction Chromatography (HIC):
Exploits hydrophobic patches on protein surfaces. Proteins bind to mildly hydrophobic matrices at high salt and are eluted as salt is lowered. - Affinity Chromatography:
Provides the highest specificity. Proteins bind to a ligand that mimics a substrate, inhibitor, or antibody, then are gently eluted to yield highly pure material. - Reversed-Phase High-Pressure Chromatography (HPLC):
Particularly effective for small peptides. It uses hydrophobic stationary phases under high pressure to achieve high resolution and speed.

Sequencing Proteins and Peptides
- Sanger Was the First to Determine the Sequence of a Polypeptide:
Frederick Sanger’s work on insulin established the first complete amino acid sequence of a protein, proving that proteins have defined primary structures. - The Edman Reaction Enables Peptides & Proteins to Be Sequenced:
Edman degradation sequentially removes one residue at a time from the amino terminus, which is then identified chromatographically. This is standard for sequencing small peptides. - Large Polypeptides Are First Cleaved into Smaller Segments:
Proteins exceeding ~50 residues are enzymatically or chemically cleaved (e.g., with trypsin, chymotrypsin, CNBr) to produce manageable fragments that can be sequenced individually and then overlapped.

Modern Advances in Primary Structure Determination
- Molecular Biology Has Revolutionized the Determination of Primary Structure:
Today, the nucleotide sequence of a gene can be translated computationally into the amino acid sequence of its encoded protein, bypassing direct chemical sequencing in many cases. - Mass Spectrometry Detects Covalent Modifications:
High-resolution MS identifies masses of peptides with extraordinary accuracy. Post-translational modifications such as phosphorylation or glycosylation are detected directly. - Tandem Mass Spectrometry (MS/MS):
Peptides are fragmented inside the instrument; the pattern of fragment ions is used to deduce sequence rapidly and sensitively. - Genomics Enables Proteins to Be Identified from Small Amounts of Sequence Data:
Even short peptide “fingerprints” can be matched to predicted proteins from DNA databases, greatly accelerating identification.
Proteomics and the Proteome
- The Goal of Proteomics:
To identify and characterize the entire complement of proteins expressed by a cell, tissue, or organism under defined conditions, including their modifications and interactions. - Two-Dimensional Electrophoresis & Gene Array Chips Are Used to Survey Protein Expression:
2-D gels separate proteins first by pI (IEF) and then by molecular weight (SDS–PAGE), producing characteristic “spots.” Coupled with mass spectrometry and gene arrays, researchers can profile expression patterns on a large scale. - Bioinformatics Assists Identification of Protein Functions:
Computational tools analyze sequence data, predict structure and domains, and compare proteins across species to infer function. Integrating experimental proteomics with bioinformatics is central to systems biology.
SUMMARY
- Long amino acid polymers or polypeptides constitute
the basic structural unit of proteins, and the structure
of a protein provides insight into how it fulfills its
functions. - The Edman reaction enabled amino acid sequence
analysis to be automated. Mass spectrometry provides a sensitive and versatile tool for determining
primary structure and for the identification of posttranslational modifications. - DNA cloning and molecular biology coupled with
protein chemistry provide a hybrid approach that
greatly increases the speed and efficiency for determination of primary structures of proteins. - Genomics—the analysis of the entire oligonucleotide
sequence of an organism’s complete genetic material—has provided further enhancements. - Computer algorithms facilitate identification of the
open reading frames that encode a given protein by
using partial sequences and peptide mass profiling to
search sequence databases.