Microbiome sequencing is a pivotal technique for analyzing the diverse communities of microorganisms—such as bacteria, archaea, fungi, and viruses—that inhabit various environments, including the human body. This process enables researchers to explore the composition, interactions, and functions of these microbial communities, thereby enhancing our understanding of their roles in health and disease.
Key Objectives in Microbial Community Research
Microbial community research focuses on several primary objectives, including:
- Species Composition and Distribution: The species composition and distribution of microbial communities in a given environment are shaped by a range of factors, such as environmental conditions and interspecies interactions. This diversity is essential in understanding community structure, and it often varies based on the environmental type and specific research goals.
- Environmental Factors Affecting Microbial Communities: Environmental variables such as temperature, pH, moisture, and nutrient availability significantly influence microbial communities. Microbes adapt to these environmental factors, and symbiotic relationships between microbial populations and their hosts can further modify distribution and ecosystem roles.
- Functional Roles of Microbial Communities: Microbial communities perform various beneficial functions, including nutrient cycling, organic matter decomposition, and nitrogen fixation. However, some microorganisms also exhibit pathogenic properties that can adversely affect plants, animals, and even humans, emphasizing the need to study both beneficial and harmful microbial species.
- Species Variability: Microbial communities display high variability in species composition. Some species may be dominant, contributing to community differences across samples, while the presence of potential pathogenic species, especially in health-related studies, is crucial in understanding ecological and health-related variations.
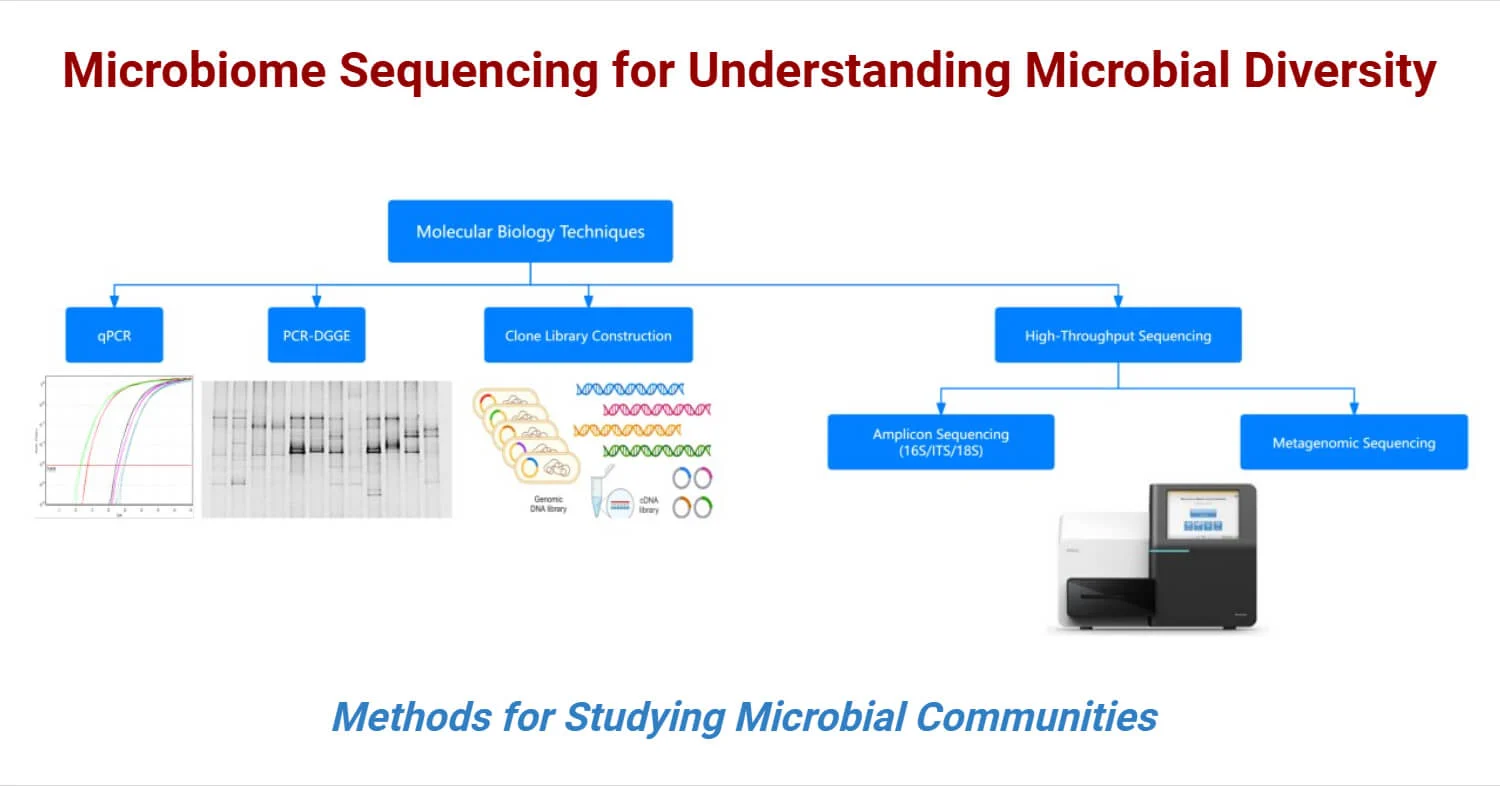
Methods for Studying Microbial Communities
Several methodologies are employed to explore microbial community diversity, structure, and functionality. These methods help determine the relative abundance of microbes and their functional potential:

- Quantitative Real-Time PCR (qPCR): qPCR is a sensitive technique that quantifies microbial species by measuring DNA amplification during the reaction. It uses specific primers and fluorescent probes to track the amplification process, allowing researchers to accurately assess microbial abundance in environmental samples. qPCR is particularly useful for quantitative microbial ecology.
- PCR-Denaturing Gradient Gel Electrophoresis (PCR-DGGE): PCR-DGGE is used to analyze microbial community diversity by separating DNA fragments based on their melting behavior in a denaturing gradient gel. The distinct banding patterns reflect variations in microbial community composition, providing insight into species diversity and community structure.
- Clone Library Construction: Clone library construction involves cloning and sequencing specific microbial genes. It helps in the analysis of gene diversity and distribution within a community. Targeted gene sequences are amplified, cloned, and sequenced to examine the functional roles of individual microorganisms within the microbial community.
- High-Throughput Sequencing (HTS): HTS methods, which include amplicon sequencing and metagenomic sequencing, offer comprehensive and cost-effective ways to analyze microbial communities. These techniques enable researchers to study microbial taxonomy, functional potential, and ecological interactions at a broad scale.

Microbiome Sequencing Methods
Microbiome sequencing techniques can be broadly categorized into amplicon sequencing and metagenomic sequencing, each with its own strengths and limitations.
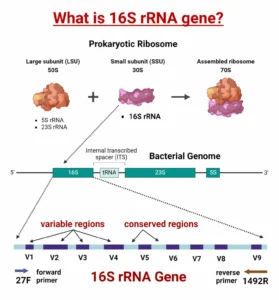
- Amplicon Sequencing: Amplicon sequencing targets specific marker genes to analyze microbial composition. Commonly targeted genes include the bacterial 16S rRNA gene, fungal ITS region, and eukaryotic 18S rRNA gene. This method is fast, cost-effective, and well-suited for studies focused on microbial diversity.Advantages of Amplicon Sequencing:
- Quick sample preparation and analysis.
- Cost-effective for large sample sizes.
- Provides broad taxonomic information.
- Suitable for studies on microbial diversity in various environmental and host-associated samples.
- Amplification bias can impact quantitative accuracy.
- Species-level resolution is often limited to the genus level.
- Functional information is restricted.
- Metagenomic Sequencing: Metagenomic sequencing captures the entire genetic material within a sample, providing detailed insights into microbial communities’ structure, function, and ecological interactions. It is capable of detecting both known and unknown microorganisms, including bacteria, fungi, viruses, and small eukaryotes.Advantages of Metagenomic Sequencing:
- Offers comprehensive data on microbial taxa and their functional genes.
- Provides species and strain-level resolution.
- Identifies previously unknown microorganisms.
- Minimizes PCR bias associated with targeted sequencing.
- More expensive and time-consuming than amplicon sequencing.
- Complex sample preparation and analysis.
- High sequencing depth required.
- May include host and organelle DNA contamination.
Choosing the Right Sequencing Method
The choice between amplicon sequencing and metagenomic sequencing depends on the research objectives:
- For Microbial Community Diversity and Composition: If the primary goal is to understand microbial diversity and composition without focusing on species-level identification, amplicon sequencing is a reliable and cost-effective method. It is particularly useful for large-scale studies where quick results are needed.
- For Comprehensive Functional and Taxonomic Analysis: Metagenomic sequencing is ideal for studies that require detailed insights into the functional potential of microbial communities. This method is essential when studying complex environments, where a thorough understanding of microbial interactions, metabolic pathways, and ecological roles is necessary.
Combining Amplicon and Metagenomic Sequencing
In some research settings, combining amplicon and metagenomic sequencing is a highly effective approach. Researchers can first use amplicon sequencing for initial screening of microbial communities, which is fast and inexpensive. Subsequently, metagenomic sequencing can be applied to selected samples to gain more detailed information about microbial diversity, species identification, and functional analysis.
This combination of methods allows for efficient use of resources while providing deep, high-resolution data on microbial communities.
CD Genomics Microbiome Sequencing Solutions
CD Genomics offers advanced microbiome sequencing services that include both amplicon and metagenomic sequencing, supporting research across various fields, including infectious diseases, agriculture, and environmental science. Their services include:
- Microbial Diversity Analysis: Using 16S/18S/ITS marker gene sequencing, researchers can examine the diversity and relative abundance of microbial groups.
- Functional Microbial Sequencing: Focuses on the functional genomes of specific microbial groups, offering insights into functional roles.
- Metagenomics: Provides a comprehensive analysis of the genetic structure of microbial communities.
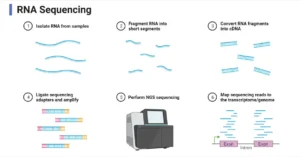
- Metatranscriptomics: RNA sequencing to study gene expression in response to environmental conditions.
By utilizing advanced sequencing platforms like NGS, PacBio SMRT, and Nanopore, CD Genomics ensures high-quality data and extensive bioinformatics support, facilitating breakthroughs in microbial research.
Conclusion
Microbiome sequencing has become an indispensable tool for studying microbial communities and their functional roles across diverse environments. By utilizing methods such as amplicon and metagenomic sequencing, researchers can uncover detailed insights into microbial diversity, interactions, and ecological functions. Whether through initial community screening or in-depth functional analysis, microbiome sequencing continues to advance our understanding of the microbial world, contributing to fields like health, agriculture, and environmental science.