DNA Replication Inhibitors (Prokaryotes)
DNA replication inhibitors in prokaryotes are compounds or drugs that disrupt the process of DNA replication in bacteria. These inhibitors have applications in both research and the development of antibiotics. Here is an exhaustive list of DNA replication inhibitors in prokaryotes along with their mechanisms of action:
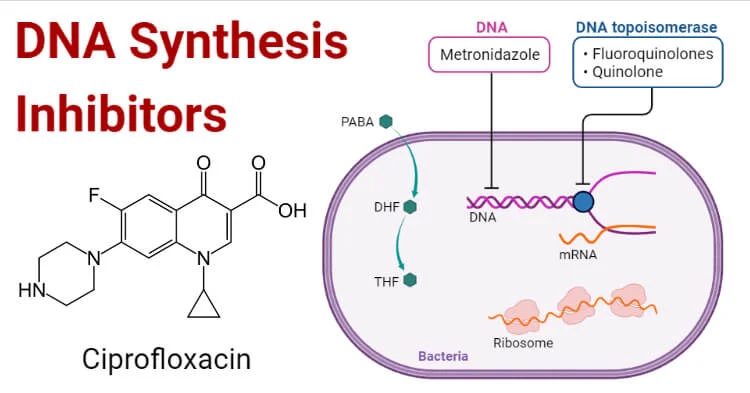
- Ciprofloxacin: A fluoroquinolone antibiotic, ciprofloxacin inhibits DNA replication by targeting DNA gyrase, an enzyme that introduces negative supercoils into DNA. Inhibition of DNA gyrase leads to the accumulation of positive supercoils and DNA strand breaks.
- Novobiocin: Novobiocin is a coumarin antibiotic that inhibits DNA replication in prokaryotes by targeting DNA gyrase. It prevents the enzyme from introducing negative supercoils into DNA, leading to problems in DNA structure and replication.
- Quinolones (e.g., nalidixic acid): Quinolones target DNA gyrase and topoisomerase IV, enzymes involved in DNA replication and repair. Inhibition of these enzymes leads to DNA damage and prevents replication.
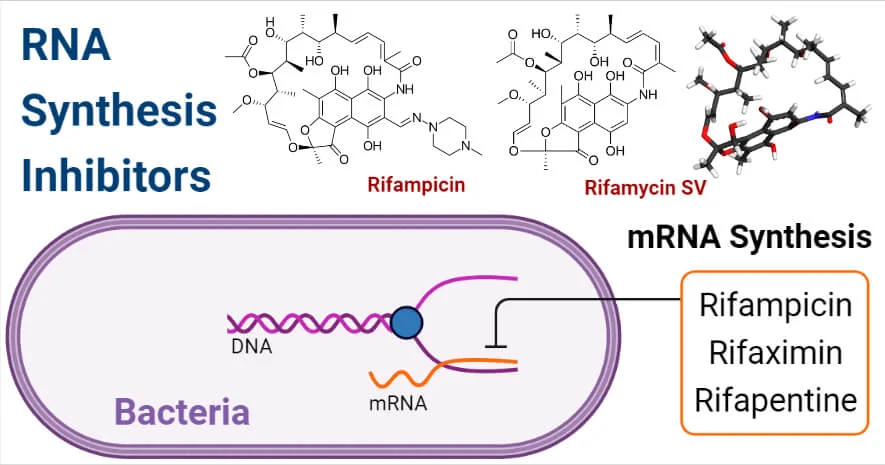
- Rifampicin: An antibiotic primarily used against tuberculosis, rifampicin inhibits DNA replication by binding to the ẞ- subunit of bacterial RNA polymerase, blocking the initiation of transcription and indirectly affecting replication.
- Sulphonamides (e.g., sulfamethoxazole): These compounds inhibit dihydropteroate synthase, an enzyme involved in folate biosynthesis. Folate is essential for DNA replication, and inhibition of this enzyme disrupts DNA synthesis.
- Trimethoprim: It inhibits dihydrofolate reductase, an enzyme in the folate biosynthesis pathway. By blocking the production of thymidine, a DNA building block, trimethoprim interferes with DNA replication.
- Metronidazole: Metronidazole disrupts DNA replication by being activated by the bacterial cell and interfering with the DNA helicase and DNA strand separation.
- Fluorouracil (5-FU): This nucleotide analog is incorporated into bacterial DNA during replication, leading to chain termination and inhibiting further DNA synthesis.
- Tetracyclines (e.g., doxycycline): These antibiotics inhibit protein synthesis in prokaryotes, affecting DNA replication indirectly by preventing the production of essential proteins.
- Actinomycin D: Actinomycin D intercalates into DNA, preventing RNA polymerase from moving along the DNA template during transcription, thereby inhibiting DNA replication.
- Nalidixic Acid: Nalidixic acid is a quinolone antibiotic that targets DNA gyrase, leading to the accumulation of DNA breaks and inhibiting replication.
- Rifabutin: A rifamycin antibiotic, rifabutin inhibits DNA replication in prokaryotes by binding to RNA polymerase and blocking transcription initiation, which indirectly affects replication.
These DNA replication inhibitors in prokaryotes act through various mechanisms, including interference with DNA gyrase and topoisomerases, inhibition of enzymes in nucleotide synthesis, interference with DNA structure, and disruption of protein synthesis. They are used for antibiotic development, microbiology research, and clinical applications.
DNA Replication Inhibitors (Eukaryotes)
DNA replication inhibitors in eukaryotes are compounds or drugs that interfere with the process of DNA replication, which is essential for cell division and genetic stability. These inhibitors are important for research and have therapeutic applications in cancer treatment and antiviral drugs. Here is an exhaustive list of DNA replication inhibitors in eukaryotes along with their mechanisms of action:
- Aphidicolin: This compound inhibits DNA replication by blocking the activity of DNA polymerases, specifically DNA polymerase a and δ.
- Hydroxyurea: It disrupts DNA replication by inhibiting ribonucleotide reductase, which is necessary for the production of deoxyribonucleotides, the building blocks of DNA.
- Cytarabine (Cytosine Arabinoside): Cytarabine is a nucleoside analog that is incorporated into DNA during replication, leading to chain termination and inhibiting further replication.
- Gemcitabine: Similar to cytarabine, gemcitabine is a nucleoside analog that interferes with DNA replication by causing chain termination.
- Bleomycin: This antibiotic inhibits DNA replication by causing DNA breaks and disrupting DNA structure.
- Methotrexate: Methotrexate is an antimetabolite that inhibits dihydrofolate reductase, a key enzyme in the biosynthesis of thymidine, a DNA building block.
- Idoxuridine: Idoxuridine is a nucleoside analog that interferes with DNA replication by replacing thymidine in DNA strands, leading to chain termination.
- Ara-C (Arabinofuranosyl Cytidine): * Ara-C, Ara-C, a nucleoside analog, is incorporated into DNA, leading to chain termination during replication.
- Adriamycin (Doxorubicin): This drug inhibits DNA replication by intercalating into DNA and causing DNA breaks.
- Camptothecin: It is a topoisomerase I inhibitor that disrupts DNA replication by causing DNA breaks and trapping the topoisomerase I-DNA complex.
- Etoposide: Similar to camptothecin, etoposide inhibits DNA replication by targeting topoisomerase II and disrupting DNA structure.
- Mitomycin C: This antibiotic inhibits DNA replication by cross-linking DNA strands, preventing DNA strand separation during replication.
- NU1025: A specific inhibitor of the enzyme poly(ADP-ribose) polymerase (PARP), which is involved in DNA repair. Inhibition of PARP interferes with the repair of DNA damage during replication.
- 3-Aminobenzamide (3-AB): Another PARP inhibitor, 3-AB interferes with DNA repair processes during replication.
- Apurinic/Apyrimidinic (AP) Site Inhibitors: Compounds like CRT0044876 inhibit the repair of AP sites, which can accumulate during DNA replication.
- DNA Ligase Inhibitors: Compounds like L189 inhibit DNA ligase, an enzyme necessary for sealing nicks in DNA strands during replication.
- Pyrazofurin: This antimetabolite inhibits orotidylate decarboxylase, which is required for the synthesis of pyrimidine nucleotides during DNA replication.
These DNA replication inhibitors act through various mechanisms, including interference with DNA polymerases, incorporation into DNA strands, disruption of DNA structure, inhibition of enzymes involved in nucleotide synthesis, and interference with DNA repair processes. They are used in both research and clinical applications to study DNA replication and as chemotherapeutic agents for various diseases, particularly cancer.
Transcription inhibitors (Prokaryotes)
Transcription inhibitors in prokaryotes are compounds that interfere with the process of transcription, which is crucial for gene expression. Here is an exhaustive list of transcription inhibitors in prokaryotes along with their mechanisms of action:
- Rifampicin: This antibiotic inhibits bacterial RNA polymerase by binding to the ẞ-subunit, preventing the initiation of RNA synthesis. It specifically hinders the formation of the first phosphodiester bond in the growing RNA chain.
- Actinomycin D: It intercalates into DNA, blocking RNA polymerase from moving along the DNA template during transcription, thereby inhibiting transcription.
- Streptolydigin: This antibiotic binds to the secondary channel of the bacterial RNA polymerase and interferes with the elongation phase of transcription, inhibiting further RNA synthesis.
- GreA and GreB proteins: These are bacterial transcription elongation factors that stimulate the release of RNA polymerase from the template, effectively terminating transcription.
- T7 lysozyme: It inhibits transcription in T7 bacteriophage by binding to T7 RNA polymerase and blocking transcription initiation.
- Tagetitoxin: This toxin inhibits transcription by binding to RNA polymerase in Escherichia coli, preventing the escape of the transcription complex from the promoter.
- Acriflavine: It interferes with RNA polymerase activity by binding to the DNA template and impeding the movement of RNA polymerase during transcription.
Transcription inhibitors (Eukaryotes)
Transcription inhibitors in eukaryotes are compounds or drugs that interfere with the process of transcription, where DNA is used as a template to produce RNA. These inhibitors are essential for both research purposes and therapeutic applications. Here is an exhaustive list of transcription inhibitors in eukaryotes along with their mechanisms of action:
- Actinomycin D: This antibiotic intercalates into DNA, preventing RNA polymerase from moving along the DNA template during transcription, thereby inhibiting transcription.
- a-Amanitin: A toxic compound found in some Amanita mushroom species, it selectively inhibits RNA polymerase II, a key enzyme in eukaryotic transcription, by binding to the enzyme and preventing elongation.
- 5,6-dichloro-1-B-D-ribofuranosylbenzimidazole (DRB): DRB inhibits RNA polymerase II by interfering with the serine 2 phosphorylation of the C-terminal domain of the polymerase, thereby blocking transcription elongation.
- Triptolide: Derived from the Chinese herb Tripterygium wilfordii, triptolide inhibits RNA polymerase II and promotes its degradation, ultimately preventing transcription.
- Thiolutin: This antibiotic inhibits RNA polymerase by binding to the enzyme and blocking transcription elongation in both prokaryotes and eukaryotes.
- Cordycepin (3′-deoxyadenosine): Cordycepin is a modified nucleoside analog that can be incorporated into RNA during transcription, leading to premature transcription termination.
- 5-Fluorouracil (5-FU): A nucleotide analog that can be incorporated into RNA during transcription, introducing faulty nucleotides and disrupting transcription.
- Actinomycin C: Actinomycin C binds to DNA and disrupts RNA polymerase movement along the DNA template, leading to transcription inhibition.
- Flavopiridol: A cyclin-dependent kinase (CDK) inhibitor, flavopiridol inhibits the phosphorylation of RNA polymerase II’s C-terminal domain, interfering with transcription elongation.
- Mithramycin A: This antibiotic binds to GC-rich regions in DNA, particularly to the GC box in promoters, and interferes with transcription initiation by inhibiting the binding of transcription factors.
- JQ1: A bromodomain inhibitor targeting BRD4, a regulator of transcription. JQ1 disrupts BRD4 functions, influencing transcriptional regulation.
- Vorinostat (SAHA): An HDAC inhibitor, vorinostat affects gene expression by altering histone acetylation patterns, allowing or preventing transcription.
- Guanidines and Amidines: Compounds like DB75 and 9AA inhibit transcription elongation by interfering with nucleotide addition.
- Mycophenolic Acid (MPA): MPA inhibits transcription by preventing inosine monophosphate (IMP) dehydrogenase activity, leading to a depletion of GTP, a necessary substrate for RNA synthesis.
- Camptothecin: It is a topoisomerase I inhibitor, which indirectly affects transcription by causing DNA breaks and interfering with DNA supercoiling, subsequently affecting RNA polymerase movement along the DNA template.
- 5-Azacytidine: A DNA methyltransferase inhibitor, affecting DNA methylation patterns and thereby indirectly influencing transcription.
These transcription inhibitors act through various mechanisms, including interference with RNA polymerase activity, elongation, DNA binding, chromatin modification, and the regulation of RNA structure. Researchers use these inhibitors in both basic science and applied research to study transcription processes and gene regulation, and they have therapeutic potential in treating diseases related to abnormal gene expression.
Translation inhibitors (Prokaryotes)
Translation inhibitors in prokaryotes are compounds or drugs that interfere with the process of translation, where the genetic code in mRNA is used to synthesize proteins. These inhibitors are crucial for research and have antibiotic applications. Here is an exhaustive list of translation inhibitors in prokaryotes along with their mechanisms of action:
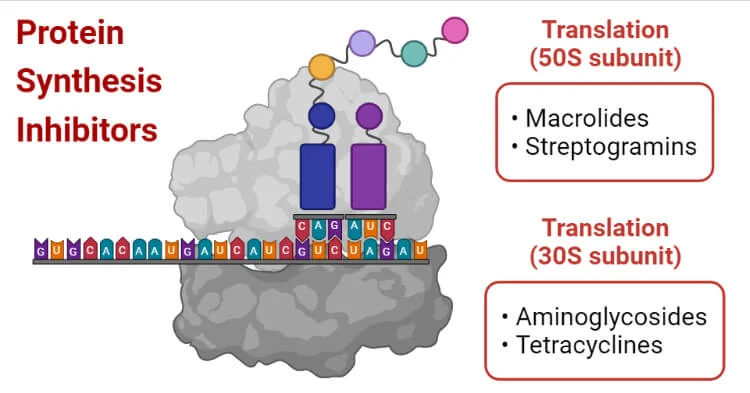
- Aminoglycosides (e.g., streptomycin, kanamycin, neomycin): These antibiotics bind to the 30S ribosomal subunit, leading to misreading of the mRNA and the incorporation of incorrect amino acids into the growing peptide chain, ultimately inhibiting translation.
- Tetracyclines (e.g., doxycycline): Tetracyclines bind to the 30S ribosomal subunit, preventing the binding of aminoacyl-tRNA to the ribosome, thereby inhibiting protein synthesis.
- Chloramphenicol: Chloramphenicol inhibits translation by binding to the 50S ribosomal subunit, inhibiting the peptidyl transferase activity responsible for peptide bond formation.
- Macrolides (e.g., erythromycin, azithromycin): These antibiotics bind to the 50S ribosomal subunit and block the exit tunnel, preventing the elongation of the peptide chain.
- Lincosamides (e.g., clindamycin): Lincosamides inhibit protein synthesis by binding to the 50S ribosomal subunit and blocking the peptidyl transferase reaction.
- Streptogramins (e.g., quinupristin-dalfopristin): Streptogramins inhibit translation by binding to the 50S ribosomal subunit, leading to the premature release e release of incomplete peptides.
- Fusidic Acid: Fusidic acid inhibits translation by preventing the release of elongation factor G (EF-G) from the ribosome, blocking translation elongation.
- Purine Analog Inhibitors (e.g., puromycin): Puromycin is an analog of aminoacyl-tRNA that gets incorporated into the growing peptide chain, leading to premature chain termination and inhibiting translation.
- Oxazolidinones (e.g., linezolid): Oxazolidinones inhibit translation by binding to the 50S ribosomal subunit and blocking the formation of the initiation complex.
- Mupirocin: Mupirocin inhibits translation by blocking the aminoacyl-tRNA synthetase, preventing the charging of tRNA with amino acids and inhibiting protein synthesis.
- Fusidic Acid: It inhibits translation by interfering with the release of EF-G from the ribosome, preventing translation elongation.
- P-P-P-Aminobenzonitrile (PPAN): PPAN inhibits translation by interfering with peptide bond formation on the ribosome.
- Kasugamycin: Kasugamycin inhibits translation by preventing the binding of initiation factor 3 (IF3) to the 30S ribosomal subunit, disrupting the translation initiation complex.
- Spectinomycin: This antibiotic inhibits translation by binding to the 30S ribosomal subunit and disrupting the binding of tRNA to the ribosome.
- Tigecycline: Tigecycline is a glycylcycline antibiotic that inhibits translation by binding to the 30S ribosomal subunit and preventing the entry of aminoacyl-tRNA.
- Retapamulin: This antibiotic inhibits translation by binding to the 50S ribosomal subunit and interfering with the formation of the elongation complex.
These translation inhibitors in prokaryotes act through various mechanisms, including interference with ribosomal subunits, blocking peptide bond formation, disrupting the binding of tRNA to the ribosome, and affecting initiation and elongation factors. They are used for antibiotic development, microbiology research, and clinical applications. the ribosome, and affecting initiation and elongation factors. They are used for antibiotic development, microbiology research, and clinical applications.
Translation inhibitors (Eukaryotes)
Translation inhibitors in eukaryotes are compounds or drugs that interfere with the process of translation, where mRNA is used to synthesize proteins. These inhibitors are essential for research and have therapeutic applications. Here is an exhaustive list of translation inhibitors in eukaryotes along with their mechanisms of action:
- Cycloheximide: This compound inhibits eukaryotic translation by blocking the translocation step of elongation. It binds to the ribosome and prevents the movement of the ribosome along the mRNA.
- Puromycin: Puromycin is an analog of aminoacyl-tRNA that gets incorporated into the growing peptide chain. This leads to premature chain termination and inhibits translation.
- Anisomycin: Anisomycin inhibits eukaryotic translation by preventing the peptidyl transferase reaction on the ribosome, blocking peptide bond formation.
- Homoharringtonine (Omacetaxine mepesuccinate): It inhibits translation initiation in eukaryotes by preventing the binding of the initiator tRNA to the ribosome.
- Harringtonine: Harringtonine, like homoharringtonine, inhibits translation initiation by preventing the formation of the initiation complex.
- Gentamicin: Gentamicin is an aminoglycoside antibiotic that can inhibit translation in eukaryotes by binding to the ribosome and causing misreading of the mRNA.
- Diphtheria Toxin: Diphtheria toxin inhibits eukaryotic translation by ADP-ribosylating and inactivating elongation factor 2 (EF-2), which is necessary for the translocation step in translation.
- Shiga Toxin: Similar to diphtheria toxin, Shiga toxin inhibits translation by ADP-ribosylating and inactivating EF-2.
- Ricin: Ricin inhibits eukaryotic translation by depurinating the 28S rRNA in the 60S ribosomal subunit, preventing the ribosome from functioning properly.
- Chloramphenicol: Chloramphenicol inhibits translation in eukaryotes by binding to the 505 ribosomal subunit, inhibiting the peptidyl transferase activity that forms peptide bonds.
- Azathioprine: Azathioprine is an immunosuppressive drug that inhibits translation by affecting the metabolism of purines, ultimately interfering with the production of aminoacyl-tRNAs.
- Mupirocin (pseudomonic acid): Mupirocin inhibits translation in eukaryotes by blocking aminoacyl-tRNA synthetases, preventing the charging of tRNAs with amino acids.
- Thiostrepton: This antibiotic inhibits translation by binding to the GTPase-associated center of the 50S ribosomal subunit, interfering with elongation factor binding and inhibiting translation.
- Fumagillin: Fumagillin inhibits eukaryotic translation by targeting the methionine aminopeptidase 2 (MetAP2), preventing proper protein processing.
These translation inhibitors in eukaryotes act through various mechanisms, including blocking ribosomal function, interfering with ribosomal translocation, preventing the binding of initiator tRNA, and disrupting elongation factor activity. They are used in research, pharmaceuticals, and clinical applications to study translation processes and as potential therapeutic agents.