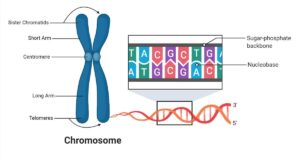
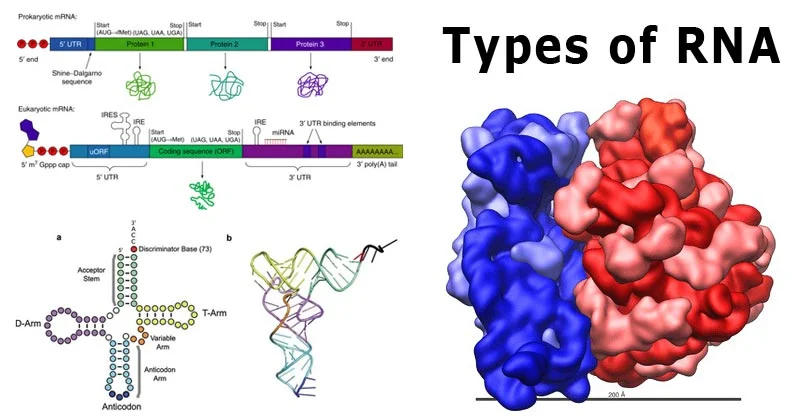
The genetic code is a fundamental aspect of molecular biology, governing how genetic information stored in DNA and transcribed into mRNA is ultimately translated into functional proteins. It is the set of rules that connects the linear sequence of nucleotides in mRNA to the linear sequence of amino acids in polypeptides. Below, we explore its defining features, patterns, and special components.

Key Characteristics of the Genetic Code
1. Triplet Nature
- The genetic code operates in groups of three nucleotides, known as codons, to specify amino acids.
- A triplet code is the minimal unit required to encode all 20 amino acids, as singlet or doublet codes provide insufficient combinations.
2. Degeneracy
- The genetic code is degenerate, meaning that multiple codons can encode the same amino acid. This feature enhances robustness against mutations.
- Partial Degeneracy: The first two bases of the codon are identical, but the third base can vary. Example: CUU and CUC both code for leucine.
- Complete Degeneracy: Any of the four bases can occupy the third position and still encode the same amino acid. Example: UCU, UCC, UCA, UCG all code for serine.
- This redundancy ensures that errors in the third nucleotide often do not alter the encoded amino acid, providing a buffer against mutations.
3. Non-Overlapping
- The genetic code is non-overlapping, meaning each nucleotide is part of only one codon.
- Adjacent codons are read sequentially, without sharing nucleotides, ensuring clarity and precision in translation.
4. Commaless
- The code is commaless, read continuously without intervening nucleotides or markers between codons.
- This uninterrupted reading ensures efficient and accurate protein synthesis.
5. Non-Ambiguity
- Each codon uniquely specifies a single amino acid, ensuring no ambiguity in translation.
- Although multiple codons may code for the same amino acid (degeneracy), no single codon codes for more than one amino acid.
6. Universality
- The genetic code is nearly universal across all forms of life, from bacteria to humans.
- Minor exceptions exist, such as in mitochondrial DNA, certain yeast species, and organisms like Mycoplasma.
- This universality underscores the shared evolutionary origin of all life.
7. Polarity
- The genetic code is read in a specific direction: 5′ → 3′.
- Reading the code in the reverse direction (3′ → 5′) would produce entirely different amino acid sequences, highlighting its inherent polarity.
Special Codons
1. Chain Initiation Codons
- AUG: Serves as the primary start codon, coding for methionine.
- GUG: Occasionally acts as a start codon in E. coli, though it typically codes for valine.
2. Chain Termination Codons
- UAA, UAG, UGA: Known as stop codons, these signal the termination of translation.
- They do not encode any amino acids but function to release the polypeptide chain from the ribosome.
3. Sense Codons
- The 61 codons that specify amino acids are referred to as sense codons.
4. Non-Sense Codons
- UAA, UAG, UGA: Initially described as non-sense codons, they are now recognized for their vital role in terminating translation and punctuating genetic messages.
Patterns in the Genetic Code
- Codons for amino acids with similar chemical properties often share structural similarities:
- Aspartic Acid (GAU, GAC) and Glutamic Acid (GAA, GAG) differ only at the third base.
- Aromatic amino acids (Phenylalanine, Tyrosine, Tryptophan) have codons that start with uracil (U).
- Codons with:
- U in the second position specify hydrophobic amino acids (e.g., Ile, Leu, Met, Phe, Val).
- A in the second position often specify charged amino acids, except arginine.
- Acidic (Asp, Glu) and basic (Arg, Lys) amino acids usually have A or G in the second position.
Reading Frames and Open Reading Frames (ORFs)
1. Reading Frames
- The mRNA sequence can be read in three possible reading frames.
- Typically, only one reading frame is functional, while the other two are interrupted by frequent stop codons.
2. Open Reading Frames (ORFs)
- An ORF is a sequence of codons starting with a start codon (AUG) and ending with a stop codon (UAA, UAG, UGA).
- Coding regions in genes contain long ORFs, whereas non-coding regions generally have shorter ORFs.
- Computational tools can analyze ORFs to predict protein sequences and identify coding regions within DNA.
Conclusion
The genetic code is an elegant and efficient system that ensures the accurate translation of genetic information into functional proteins. Its robustness, universality, and precision highlight its evolutionary significance and its central role in the molecular biology of all organisms. From the simplicity of bacterial cells to the complexity of human systems, the genetic code remains a cornerstone of life, emphasizing the interconnectedness of all living beings.