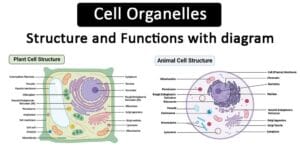
Mechanisms of Protein Targeting and Transport, Protein segregation, trafficking, and transport are critical processes within cells that ensure proteins are correctly synthesized, folded, modified, and delivered to their appropriate destinations. Here’s an overview of these processes:
Protein Targeting & Segregation
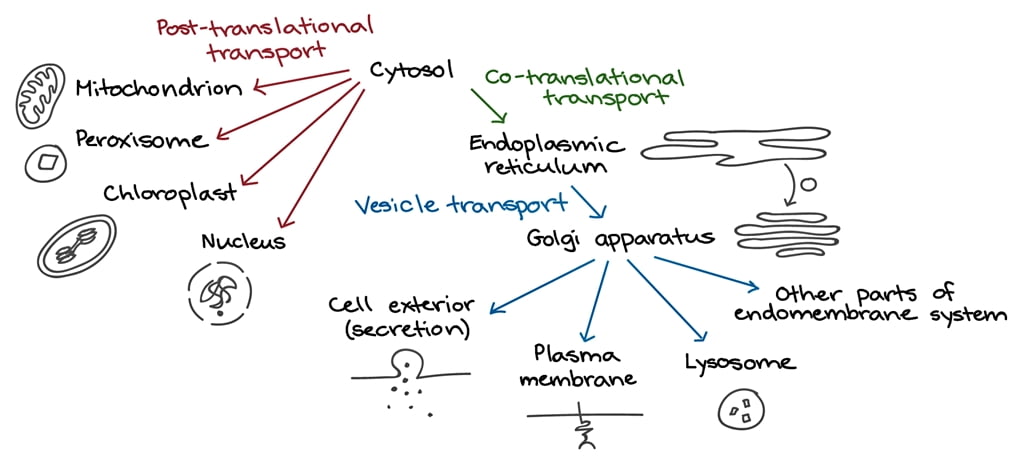
Protein segregation involves the distribution of newly synthesized proteins into different cellular compartments. This process ensures that proteins are sent to the correct organelles or cell membranes, where they perform their functions.
- Signal Peptides and Sequences: Proteins often have specific signal peptides or sequences that direct them to their destined locations. For instance, a signal sequence on a protein can direct it to the endoplasmic reticulum (ER), mitochondria, or other organelles.
- Sorting Receptors: Sorting receptors recognize these signal sequences and guide the proteins to their correct location. For example, in the ER, signal recognition particles (SRPs) bind to signal sequences and direct the ribosome-protein complex to the ER membrane.
Protein Trafficking
Protein trafficking involves the movement of proteins within the cell, especially from the site of synthesis (ribosomes) to various destinations.
- Endoplasmic Reticulum (ER): Many proteins are synthesized on ribosomes bound to the ER. These proteins enter the ER lumen, where they undergo folding and post-translational modifications.
- Golgi Apparatus: After the ER, proteins are transported to the Golgi apparatus in vesicles. The Golgi further modifies proteins and sorts them for transport to their final destinations.
- Vesicular Transport: Small vesicles bud off from the ER and Golgi, carrying proteins to various parts of the cell. These vesicles fuse with target membranes to deliver their cargo.
Protein Transport
Protein transport refers to the mechanisms by which proteins are moved across cellular membranes and within the cell.
- Cytoskeletal Elements: Motor proteins such as kinesin and dynein move vesicles along microtubules, while myosin transports vesicles along actin filaments.
- Transport Across Membranes: Proteins may be transported across membranes through translocons (e.g., the Sec61 complex in the ER membrane) or other transport channels.
- Endocytosis and Exocytosis: Endocytosis involves the internalization of proteins from the cell membrane, while exocytosis is the process of vesicles fusing with the plasma membrane to release their contents outside the cell.
Key Components and Pathways
- Endoplasmic Reticulum (ER): The ER is the starting point for the synthesis of membrane-bound and secretory proteins. The rough ER is studded with ribosomes and is involved in protein synthesis and folding.
- Golgi Apparatus: The Golgi modifies, sorts, and packages proteins received from the ER. It consists of cisternae, with distinct cis, medial, and trans regions, each responsible for different modifications.
- Lysosomes: Proteins destined for degradation are transported to lysosomes, where they are broken down by hydrolytic enzymes.
- Endosomes: These are involved in sorting endocytosed material and can direct proteins to lysosomes or back to the cell surface.
- Nucleus: Nuclear localization signals (NLS) direct proteins to the nucleus through nuclear pore complexes.
Nuclear protein transport
Nuclear protein transport is a highly regulated process that involves the selective import and export of proteins between the cytoplasm and the nucleus. This transport is critical for numerous cellular functions, including gene expression, DNA replication, and cell cycle regulation. The mechanism primarily relies on nuclear pore complexes (NPCs), signal sequences, and transport receptors.
Key Components of Nuclear Protein Transport
- Nuclear Pore Complexes (NPCs): Large multiprotein structures embedded in the nuclear envelope that act as gateways for molecular traffic between the nucleus and cytoplasm. NPCs allow the passive diffusion of small molecules and mediate the active transport of larger proteins and RNA.
- Nuclear Localization Signals (NLS): Short amino acid sequences within proteins that are destined for the nucleus. NLS sequences are recognized by nuclear transport receptors.
- Nuclear Export Signals (NES): Sequences that direct proteins out of the nucleus. NES sequences are also recognized by specific transport receptors.
- Transport Receptors (Karyopherins): These are proteins that facilitate the transport of cargo proteins through NPCs. They include importins (for nuclear import) and exportins (for nuclear export).
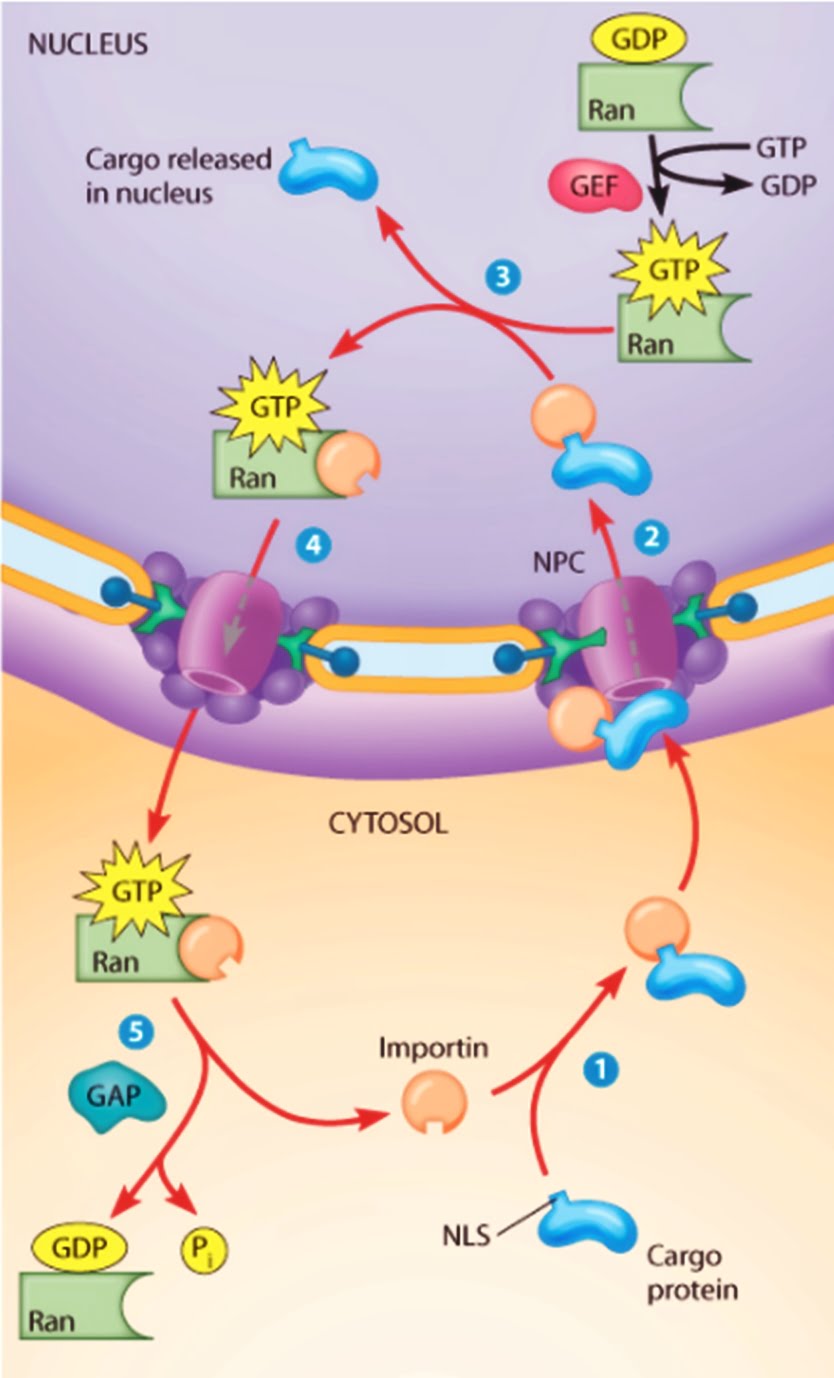
- Ran GTPase: A small GTP-binding protein that provides directionality to nuclear transport. Ran exists in two states: Ran-GTP in the nucleus and Ran-GDP in the cytoplasm.
Nuclear Import Mechanism
- Recognition: Proteins destined for the nucleus contain NLS sequences, which are recognized by importin α (an adapter protein) or directly by importin β (a transport receptor).
- Binding: The NLS-containing protein (cargo) binds to importin α, which in turn binds to importin β. Alternatively, some cargos can directly bind to importin β without the need for importin α.
- Docking: The importin-cargo complex docks at the cytoplasmic side of the NPC.
- Translocation: The complex translocates through the NPC. This process is facilitated by interactions between importins and nucleoporins (proteins that make up the NPC).
- Release: Once inside the nucleus, Ran-GTP binds to importin β, causing a conformational change that releases the cargo.
- Recycling: The importin-Ran-GTP complex is transported back to the cytoplasm, where GTP hydrolysis by Ran GTPase-activating protein (RanGAP) converts Ran-GTP to Ran-GDP, releasing importin β for another round of transport.
Nuclear Export Mechanism
- Recognition: Proteins destined for export contain NES sequences, which are recognized by exportins (such as CRM1/exportin 1).
- Binding: The NES-containing protein (cargo) binds to the exportin along with Ran-GTP, forming a trimeric complex.
- Docking: The exportin-cargo-Ran-GTP complex docks at the nuclear side of the NPC.
- Translocation: The complex translocates through the NPC to the cytoplasmic side.
- Release: In the cytoplasm, RanGAP stimulates the hydrolysis of Ran-GTP to Ran-GDP. This causes the disassembly of the complex and release of the cargo.
- Recycling: Exportin and Ran-GDP are transported back to the nucleus, where Ran nucleotide exchange factor (RanGEF) converts Ran-GDP back to Ran-GTP.
Regulation of Nuclear Transport
- Post-Translational Modifications: Phosphorylation, ubiquitination, and other modifications of cargo proteins can influence their recognition by importins or exportins.
- Interaction with Other Proteins: Binding of cargo proteins to other proteins can mask or expose NLS or NES sequences, thereby regulating their transport.
- Cell Cycle-Dependent Mechanisms: The transport machinery and NPCs can be regulated differently during various phases of the cell cycle.
Examples of Nuclear Protein Transport
- Transcription Factors: Proteins like NF-κB are transported into the nucleus in response to specific signals, where they regulate gene expression.
- Ribosomal Proteins: Synthesized in the cytoplasm and transported into the nucleus to be assembled into ribosomes.
- Histones: Imported into the nucleus to associate with DNA and form chromatin.
Understanding the detailed mechanisms of nuclear protein transport is essential for insights into cellular regulation and the pathology of diseases such as cancer, where transport processes can be disrupted.
Endoplasmic Reticulum (ER) Targeting
- Co-Translational Translocation:
- Signal Peptide: Proteins destined for the ER have an N-terminal signal peptide.
- Signal Recognition Particle (SRP): The signal peptide is recognized by the SRP, which pauses translation and directs the ribosome to the ER membrane.
- Translocon: The ribosome-SRP complex binds to the SRP receptor on the ER membrane, and the growing polypeptide is translocated into the ER lumen through the translocon (Sec61 complex).
- Signal Peptidase: The signal peptide is cleaved off by signal peptidase.
- Post-Translational Translocation:
- Proteins synthesized in the cytoplasm are targeted to the ER post-translationally with the help of chaperones and specific receptors on the ER membrane.
Mitochondrial Targeting
- Mitochondrial Targeting Sequences (MTS):
- Proteins destined for the mitochondria contain N-terminal MTS.
- TOM Complex: The translocase of the outer membrane (TOM) complex recognizes and transports these proteins across the outer membrane.
- TIM Complex: The translocase of the inner membrane (TIM) complex facilitates their movement across or into the inner membrane.
- Chaperones: Cytosolic chaperones keep the precursor proteins unfolded, while mitochondrial chaperones help in their folding once inside the mitochondria.
Chloroplast Targeting
- Chloroplast Transit Peptides (cTP):
- Proteins destined for chloroplasts have an N-terminal cTP.
- TOC and TIC Complexes: The translocon of the outer chloroplast membrane (TOC) and inner chloroplast membrane (TIC) complexes facilitate the import of these proteins.
- Stromal Processing Peptidase: cTPs are cleaved off in the stroma, and chaperones assist in folding and further targeting within the chloroplast.
Peroxisomal Targeting
- Peroxisomal Targeting Signals (PTS):
- PTS1 and PTS2: Proteins destined for peroxisomes have C-terminal (PTS1) or N-terminal (PTS2) targeting signals.
- PEX Proteins: Receptors (PEX5 for PTS1 and PEX7 for PTS2) recognize these signals and transport the proteins through the peroxisomal membrane.
Lysosomal Targeting
- Mannose-6-Phosphate (M6P) Tag:
- Lysosomal hydrolases are tagged with M6P in the Golgi apparatus.
- M6P Receptors: These receptors in the trans-Golgi network recognize and direct the tagged enzymes to lysosomes via vesicular transport.
Plasma Membrane and Secretory Pathways
- Transmembrane Proteins:
- Stop-Transfer and Signal-Anchor Sequences: These sequences halt translocation into the ER and integrate the protein into the membrane.
- Orientation: Determined by the positive-inside rule and the distribution of charged residues around the transmembrane domain.
- Secreted Proteins:
- ER-Golgi Transport: Proteins are transported from the ER to the Golgi in vesicles, undergoing further modifications.
- Golgi to Plasma Membrane: Vesicles bud off from the Golgi and fuse with the plasma membrane, releasing the protein outside the cell.
Endocytosis and Recycling Pathways
- Clathrin-Mediated Endocytosis:
- Cargo Selection: Specific receptors bind to extracellular ligands.
- Clathrin Coated Pits: These receptors cluster in clathrin-coated pits that bud off to form vesicles.
- Endosomes: The vesicles fuse with early endosomes, where the cargo is sorted.
- Recycling or Degradation: Cargo can be recycled back to the membrane or sent to lysosomes for degradation.
Golgi Targeting
- Retention Signals:
- COPI Vesicles: Retrieval of ER resident proteins is mediated by COPI vesicles, recognizing KDEL (Lys-Asp-Glu-Leu) or KKXX (Lys-Lys-X-X) motifs for retrieval back to the ER.
- Golgi Retention: Golgi resident proteins have retention signals that ensure their localization within the Golgi apparatus.
Non-Canonical Pathways
- Unconventional Protein Secretion:
- Some proteins bypass the classical ER-Golgi route and are secreted via alternative pathways, such as direct translocation across the plasma membrane or via secretory lysosomes.
These mechanisms ensure that proteins are accurately directed to their functional locations, maintaining cellular organization and efficiency. Disruptions in these pathways can lead to various diseases, highlighting their critical role in cell biology.
Quality Control Mechanisms
Cells have quality control mechanisms to ensure only properly folded and modified proteins reach their destinations.
- Chaperones: These proteins assist in the proper folding of nascent proteins and prevent aggregation.
- Unfolded Protein Response (UPR): If misfolded proteins accumulate in the ER, the UPR is activated to restore normal function by enhancing the production of chaperones and degrading misfolded proteins.
- Proteasomes: Misfolded or damaged proteins are ubiquitinated and directed to proteasomes for degradation.
Disorders Related to Protein Trafficking
Defects in protein segregation, trafficking, and transport can lead to various diseases, including:
- Cystic Fibrosis: Caused by mutations in the CFTR gene, leading to defective trafficking of the CFTR protein to the cell surface.
- Alzheimer’s Disease: Accumulation of misfolded proteins and defective trafficking can contribute to the formation of amyloid plaques.
- Lysosomal Storage Diseases: Result from defects in lysosomal enzymes, leading to the accumulation of undigested substrates.
Understanding protein segregation, trafficking, and transport is crucial for comprehending cellular function and the basis of many diseases. Research in this field continues to reveal new insights into the intricate mechanisms that govern protein dynamics within cells.