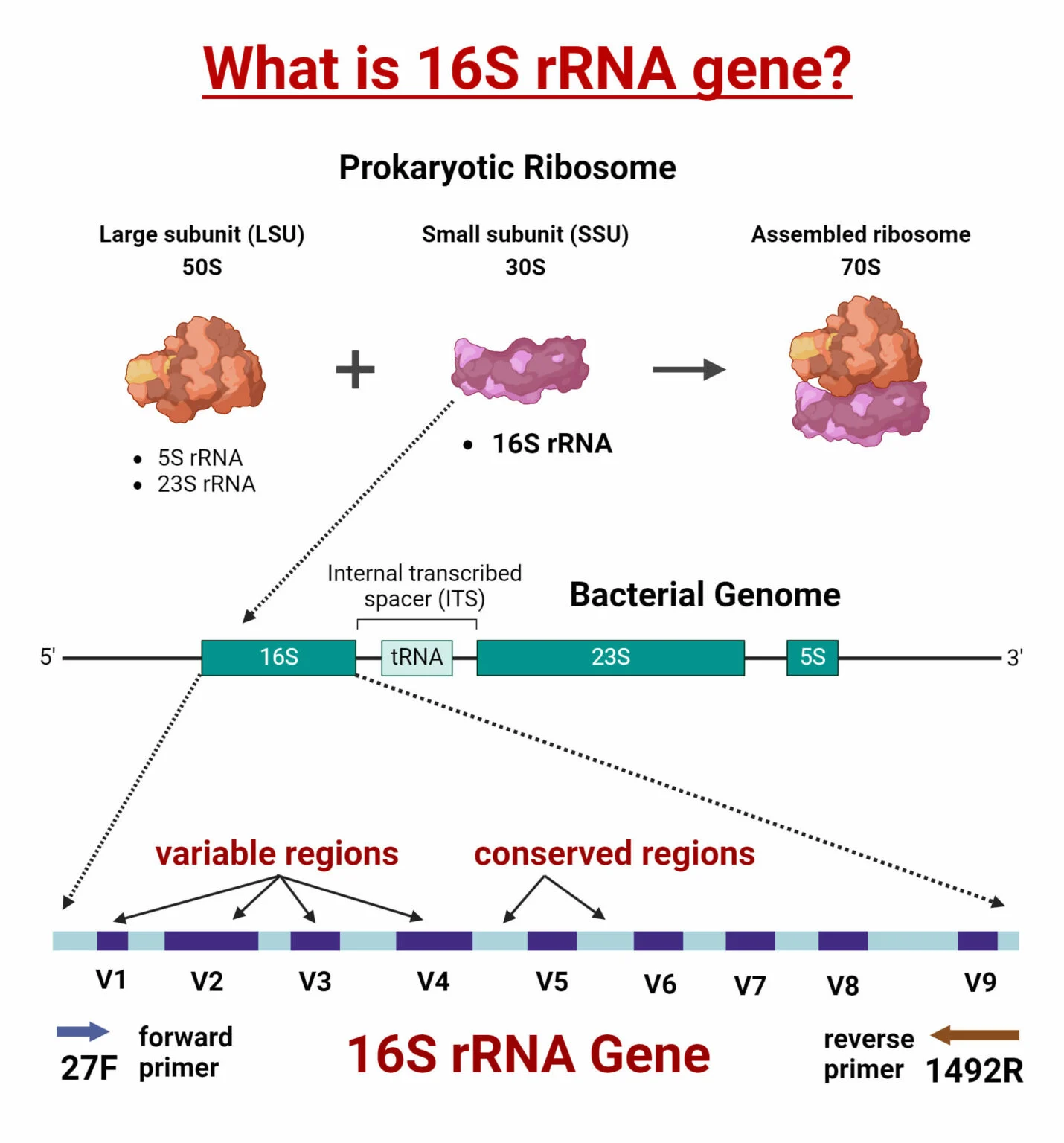
What is the 16S rRNA Gene?
The 16S rRNA gene is a widely used genetic marker for bacterial taxonomy and phylogeny. Found in the ribosomes of prokaryotes, the 16S rRNA gene is about 1500 base pairs long and contains both conserved and variable regions. The conserved regions are similar across bacterial species, making them ideal for universal amplification, while the variable regions allow differentiation and identification between species.

Principle of 16S rRNA Gene Sequencing
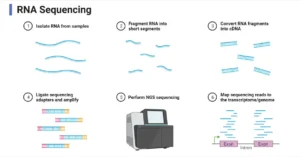
The method relies on amplicon sequencing that focuses on amplifying the 16S rRNA gene. Since the gene is highly conserved across bacteria and archaea but exhibits enough variation in its sequences to differentiate species, it serves as a powerful marker for identifying and classifying microorganisms. The process involves DNA extraction, PCR amplification of the 16S rRNA gene, sequencing, and bioinformatics analysis to identify microbial communities in samples.
Steps Involved in 16S rRNA Gene Sequencing
- Sample Collection: Environmental or biological samples (e.g., soil, water, or gut microbiome) are collected. Special care is taken to avoid contamination during the process.
- DNA Extraction: The microbial DNA is extracted using chemical and physical methods such as sonication, lysis, and purification techniques.
- PCR and Library Construction: The 16S rRNA gene is amplified using PCR, followed by library construction. DNA is fragmented, adaptors are added, and the library is purified to ensure uniformity.
- Sequencing: The DNA library undergoes high-throughput sequencing using platforms like Illumina, PacBio, or Oxford Nanopore to generate millions of short DNA sequences.
- Data Analysis: Sequencing data is processed using bioinformatics tools like QIIME or MOTHUR to analyze microbial communities. Reads are clustered into Operational Taxonomic Units (OTUs), and taxonomic identities are assigned based on reference databases.

Advantages of 16S rRNA Gene Sequencing
- Non-culturable Microbes: Unlike traditional methods, it allows the identification of microbes that cannot be cultured in a lab.
- High Throughput: Enables the sequencing of thousands of microbial DNA fragments simultaneously.
- Accurate Identification: Provides higher precision and less contamination compared to traditional culture-based methods.
- Versatile: Can be applied to diverse biological samples from different environments.
Limitations of 16S rRNA Gene Sequencing
- Resolution: It may not resolve species-level differences, especially for closely related species.
- Taxonomic Resolution: Only provides genus-level resolution in some cases.
- Data Complexity: Requires sophisticated bioinformatics tools and expertise to interpret complex data.
Applications of 16S rRNA Gene Sequencing
- Microbial Diversity Studies: Used to explore microbial diversity in soil, water, or human microbiomes.
- Clinical Microbiology: Helps identify microbial communities related to diseases, providing insights into disease mechanisms and treatment options.
- Food Safety: Identifies food-borne pathogens in fermented foods.
- Agriculture: Assists in studying the microbial communities of soil and plants, which can improve crop health.
- Human Health: Contributes to research on the human microbiome and its impacts on health, as seen in projects like the Human Microbiome Project.
Example Applications:
- Human Microbiome Project: Studies microbial communities associated with human health.
- Global Ocean Sampling Expedition: Explores microbial diversity in ocean ecosystems.
- Soil Microbiome Studies: Investigates the relationship between soil microorganisms and plant health.
- Antibiotic Resistance Studies: Tracks microbial resistance in hospital settings.
By providing detailed insights into microbial communities, 16S rRNA gene sequencing is a vital tool for studying ecology, disease, and environmental health.