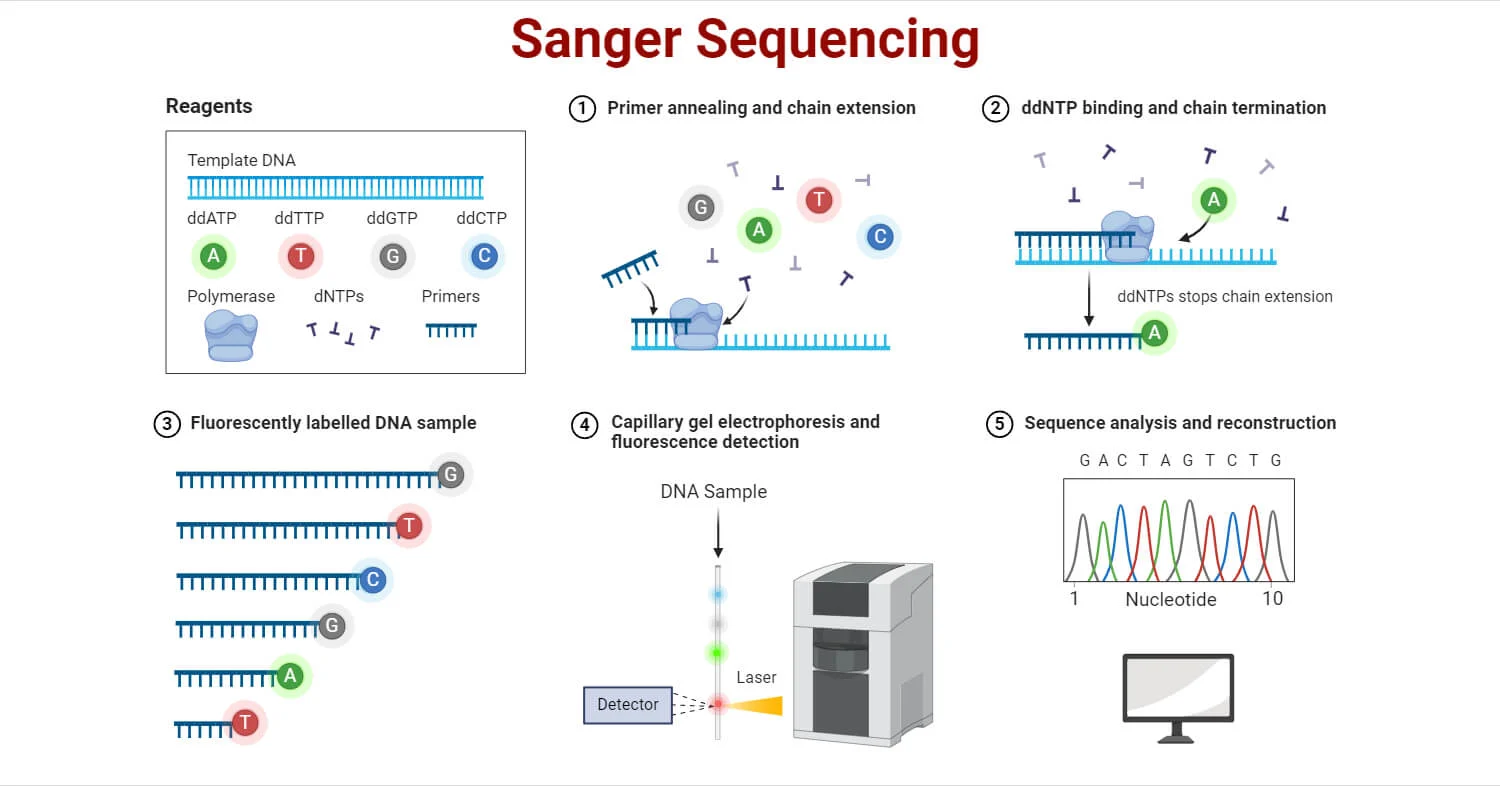
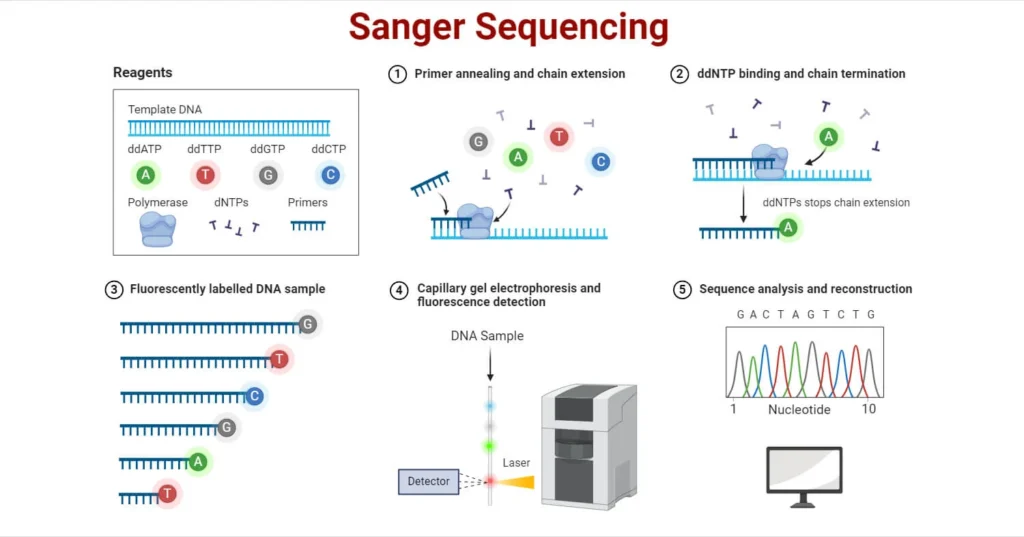
Principle of Sanger Sequencing: Sanger sequencing, also known as dideoxy sequencing or chain termination method, identifies the nucleotide sequence of DNA by incorporating modified nucleotides called dideoxynucleotide triphosphates (ddNTPs). These ddNTPs lack the 3’ hydroxyl group necessary for forming a phosphodiester bond, thereby terminating the elongation of the DNA strand. As a result, various fragments of different lengths are produced, each ending with a specific ddNTP. These fragments are then separated and analyzed to deduce the DNA sequence.
Steps Involved in Sanger Sequencing:
- DNA Template Preparation:
- DNA of interest is extracted using various methods like chemical extraction or column-based extraction.
- The extracted DNA is then converted into single-stranded molecules suitable for sequencing.
- Chain Termination PCR:
- The target DNA is amplified using Polymerase Chain Reaction (PCR), which includes template DNA, primers, dNTPs, DNA polymerase, and ddNTPs (each labeled with a distinct fluorescent marker).
- The ddNTPs terminate DNA elongation, resulting in fragments of various lengths that are terminated at different points.
- In traditional methods, four separate tubes were used for each ddNTP; in automated systems, all ddNTPs are combined in one reaction.
- Separation of DNA Fragments:
- The fragments are separated by size using electrophoresis, typically in a polyacrylamide gel or capillary electrophoresis (CE).
- In capillary electrophoresis, DNA fragments are run in a single capillary filled with gel polymer.
- Detection and Analysis:
- The separated fragments pass through a fluorescence detector, which identifies the fluorescent label attached to each ddNTP.
- The detected signals are used to generate a chromatograph, where the sequence of the DNA is determined by reading the order of the fluorescent labels.
Advantages of Sanger Sequencing:
- High accuracy, making it ideal for small-scale projects or for confirming sequences obtained by other methods.
- Suitable for sequencing short DNA regions and producing long read lengths.
- Straightforward data analysis with less complex bioinformatics tools compared to next-generation sequencing (NGS).
- Reliable and reproducible outcomes, with clear and interpretable chromatograms.
Limitations of Sanger Sequencing:
- Suitable only for small-scale projects as it is slow, expensive, and can sequence only one fragment at a time.
- Not ideal for large genomes due to high cost and complexity.
- Limited sensitivity for detecting rare mutations and complex mixtures.
- Requires high-quality, pure DNA samples for accurate results.
Applications of Sanger Sequencing:
- Medical Diagnosis: Identifying genes associated with diseases and conducting genetic testing.
- Evolutionary Studies: Identifying new species and studying evolutionary history through gene sequence comparison.
- Forensic Science: Personal identification and DNA fingerprinting in criminal cases.
- Agriculture: Identifying different breeds of crops and livestock.
- Conservation: Studying genetic diversity in species.
- Emerging Technologies: Single-cell sequencing and synthetic biology applications.
Sanger Sequencing vs. Next-Generation Sequencing (NGS):
| Characteristic | Sanger Sequencing | Next-Generation Sequencing (NGS) |
|---|---|---|
| Principle | Chain termination (dideoxy) method | Parallel sequencing (massively parallel sequencing) |
| Speed | Slower, one fragment at a time | Fast, millions of fragments simultaneously |
| Read Length | Long reads | Shorter reads |
| Cost | High for large projects, low for single genes | Economical for high-throughput projects |
| Data Analysis | Straightforward | Requires complex bioinformatics tools |
| Applications | Small-scale projects, confirmation of NGS data | Large-scale projects, whole-genome sequencing |
Sanger sequencing is often represented by a diagram showing four separate reaction tubes (or a single automated reaction) with different fluorescent ddNTPs and the subsequent separation and detection of DNA fragments by electrophoresis, leading to a chromatogram for sequence interpretation.