Helicos Single-Molecule Sequencing (tSMS) is a third-generation DNA sequencing technology that directly sequences individual DNA or RNA molecules without requiring amplification. Launched in 2008 by Helicos Biosciences, it revolutionized sequencing by minimizing preparation steps. However, the technology faced commercial challenges, leading to the company’s closure in 2012. Despite its fall, tSMS paved the way for advanced sequencing methods.
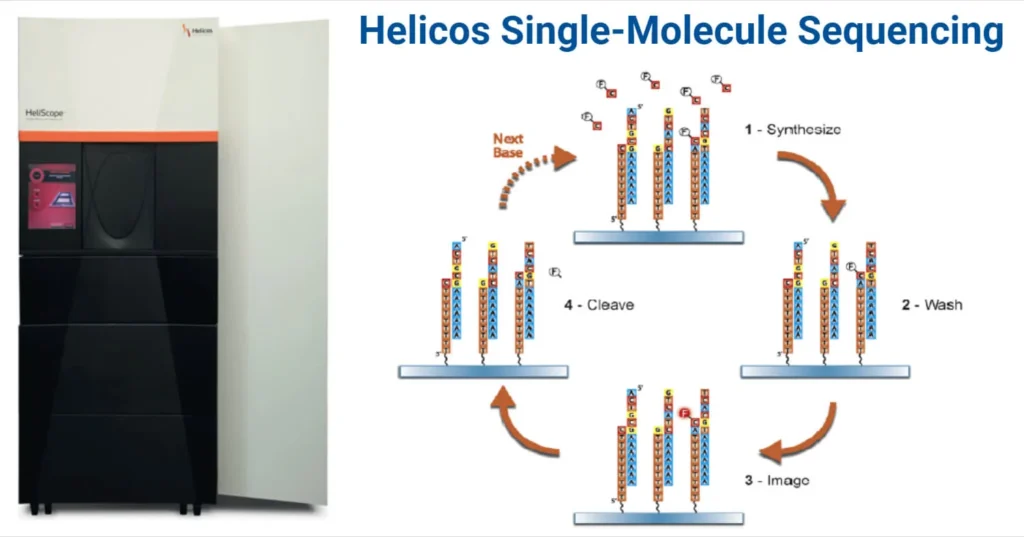
Principle of Helicos Single-Molecule Sequencing
Helicos sequencing is based on sequencing-by-synthesis (SBS) and eliminates the need for DNA amplification. Its process includes:
- Hybridization of fragmented, poly-A-tailed single-stranded DNA (ssDNA) to a glass flow cell with complementary oligo-dT primers.
- Sequential addition of fluorescently-labeled nucleotides.
- Laser-induced fluorescence detection of nucleotide incorporation.
- Iterative cycles of nucleotide addition and imaging.
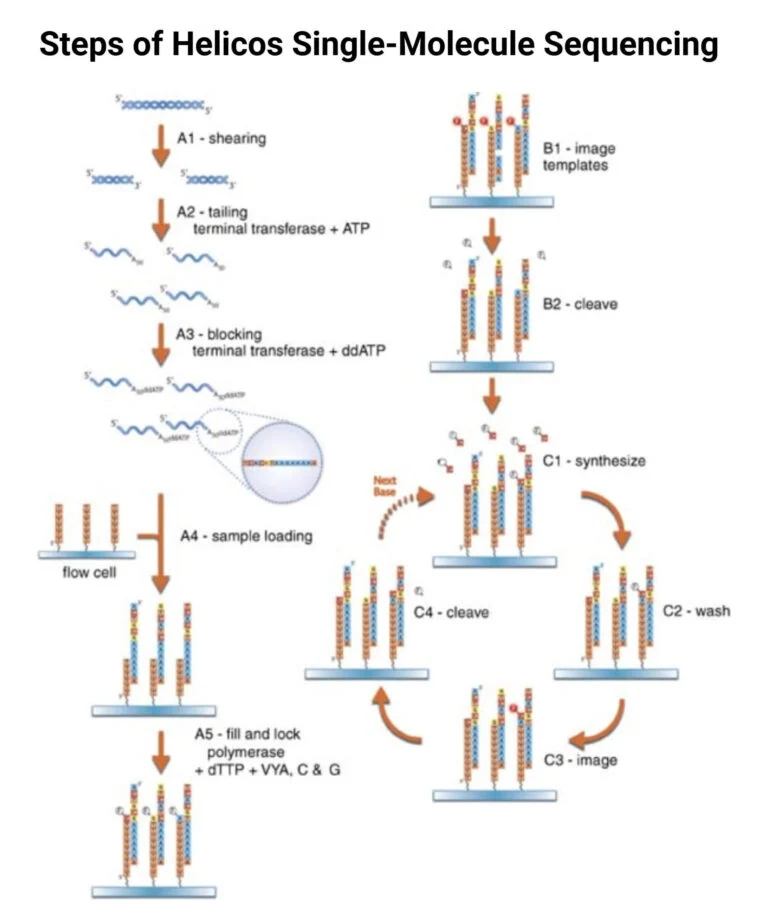
Steps of Helicos Single-Molecule Sequencing
1. Template Preparation
- Fragmentation: DNA is fragmented using sonication or enzymatic shearing.
- Denaturation: Produces single-stranded DNA (ssDNA).
- Poly-A Tailing: Facilitates attachment to flow cell primers.
- Immobilization: Tailed DNA is anchored onto a flow cell surface.
2. Fill and Lock Step
- Fill Step: dTTP and polymerase fill the poly-A tail gaps.
- Lock Step: Fluorescently-labeled terminators prevent undesired extension, preparing templates for sequencing.
3. Sequencing-by-Synthesis (SBS)
- Sequentially incorporates complementary fluorescently-labeled nucleotides.
- Fluorescence signals are captured using a CCD camera.
- Dye and terminator removal allow repeated cycles of nucleotide addition.
4. Data Analysis
- Captured images are processed using base-calling algorithms.
- The nucleotide sequences undergo downstream analysis, including genome alignment and variant detection.
Advantages of Helicos Single-Molecule Sequencing
- No Amplification: Preserves original DNA/RNA sequences, minimizing biases.
- Low Input Material: Suitable for rare or degraded samples (e.g., forensic, ancient DNA).
- Simplified Workflow: Direct hybridization reduces sample loss.
- High GC Content Tolerance: Reliable sequencing of complex genomic regions.
Limitations of Helicos Single-Molecule Sequencing
- Short Read Lengths: Limits assembly of complex genomes.
- High Costs: Expensive instruments and operational expenses.
- Environmental Sensitivity: Requires stable environments to minimize vibration-induced errors.
- Data Complexity: Generates vast data requiring advanced computational resources.
- Limited Throughput: Lagged behind competitors like Illumina in scalability and efficiency.
Applications of Helicos Single-Molecule Sequencing
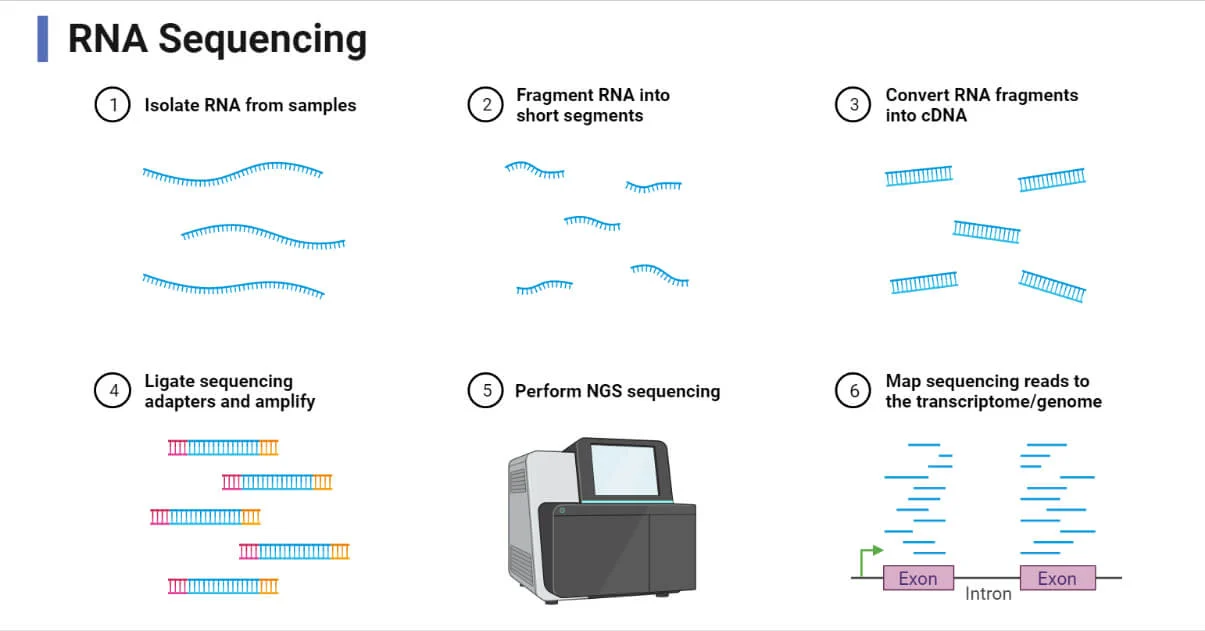
- RNA Sequencing: Direct sequencing of RNA without reverse transcription.
- ChIP-Seq: Studies regulatory and epigenetic mechanisms.
- Genetic Variant Detection: Accurate identification of rare mutations, especially in cancer research.
- Degraded DNA Analysis: Ideal for ancient or forensic DNA samples.
- High GC Content Genomes: Reliable sequencing with minimal amplification bias.
The Fall of Helicos Single-Molecule Sequencing
Helicos Biosciences faced technical and economic hurdles, such as:
- High Costs: Instruments and operations were expensive.
- Technical Challenges: Short reads, high error rates, and throughput limitations.
- Market Competition: Dominated by cost-effective and efficient platforms like Illumina.
The company ceased operations in 2011, but its foundational work inspired technologies like PacBio sequencing and Oxford Nanopore sequencing.
Legacy and Repurposing
- SeqLL: Focuses on niche applications like RNA sequencing using modified Helicos technology.
- Direct Genomics: Attempted to apply tSMS in specialized areas but faced controversy due to its founder.
Despite its commercial failure, Helicos left a legacy in single-molecule sequencing, influencing modern genomic research and technology.